| (7) B-CELL | (9) CELL-LINE | (1) CERVIX | (1) HEART | (1) LIVER | (1) OTHER | (5) SKIN |
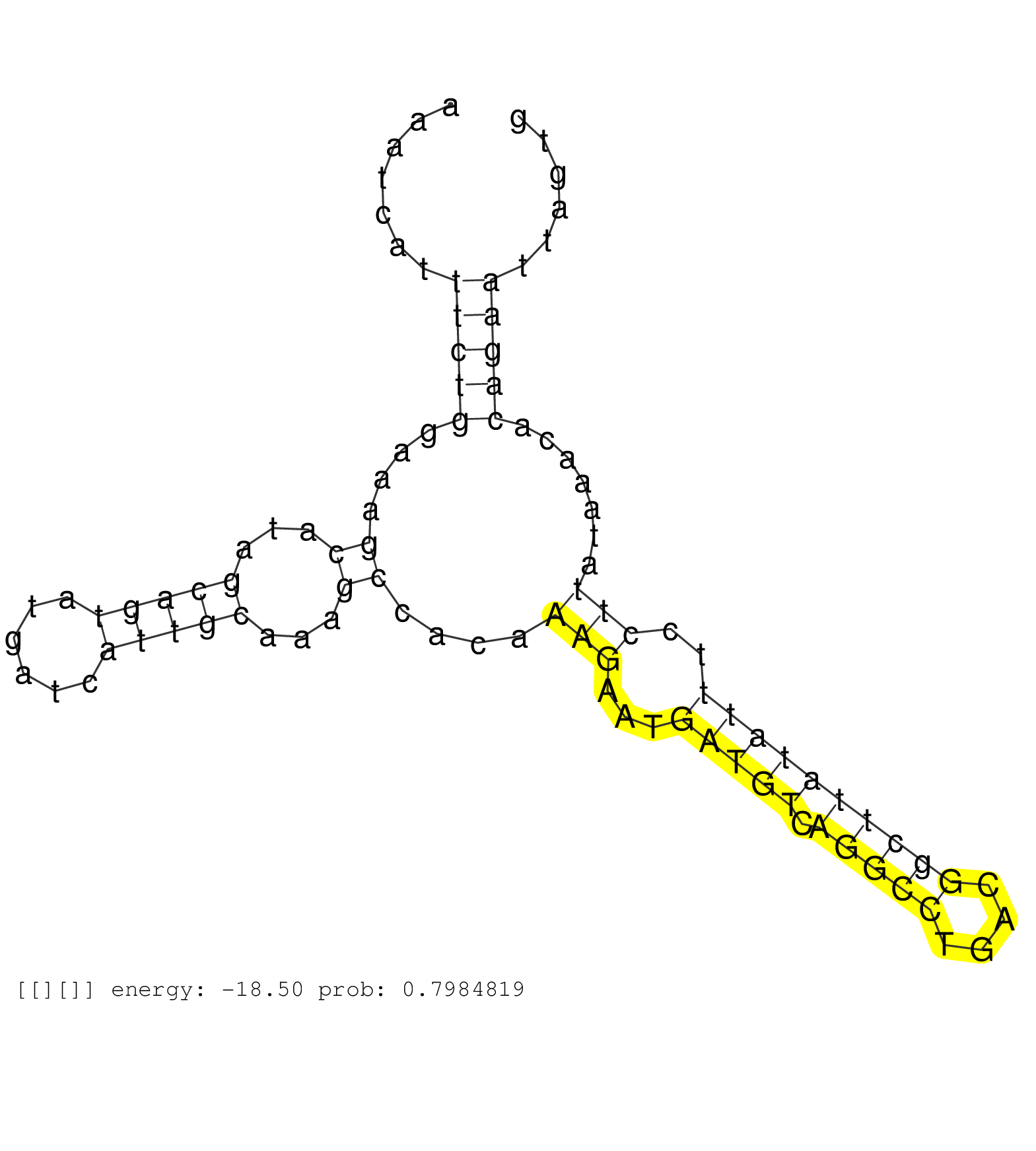
| GGTCTCCCTCACTCCAGAAGAATTTGACCAACTCCAGAAATATTCAGAATGTGAGTAAATGCATTGACAGCCCTGGGACGGGATTTCTTCTTGGCTGGAGAAATCATTTCTGGAAAGCATAGCAGTATGATCATTGCAAAGCCACAAAGAATGATGTCAGGCCTGACGGCTTATATTTCCTTATAAACACAGAATTAGTGTATTTAACATAAAAACGCAATTATCTTTAAGCTCAGTAAATACTACGTGC .................................................................................(((((((((......)))))))))..((((.....((...(((((......)))))...)).....)))).......(((((....))))).............................................................................. ................................................................................81.........................................................................................172............................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM532874(GSM532874) G699T. (cervix) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGACG.................................................................................. | 22 | 1 | 69.00 | 69.00 | 17.00 | 8.00 | 8.00 | 7.00 | 9.00 | 6.00 | - | 5.00 | 1.00 | 1.00 | 2.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGACG.................................................................................. | 23 | 1 | 28.00 | 28.00 | 4.00 | 5.00 | 7.00 | 4.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGACGG................................................................................. | 24 | 1 | 20.00 | 20.00 | 10.00 | 2.00 | - | - | - | 1.00 | 2.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGACGG................................................................................. | 23 | 1 | 14.00 | 14.00 | 8.00 | 2.00 | - | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGACGGCT............................................................................... | 26 | 1 | 6.00 | 6.00 | - | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGA.................................................................................... | 21 | 1 | 6.00 | 6.00 | - | 1.00 | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGACGGC................................................................................ | 24 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGACGGCT............................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGA.................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAAGAATTTGACCAACTCCAGAAA.................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGACGGCTTA............................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGATG.................................................................................. | 22 | 1 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGACGGC................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GTCTCCCTCACTCCAGAAGAATT.................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGACGGCTT.............................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGACGGCTTA............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGACGGA................................................................................ | 24 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGACGGCA............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAAGAATGATGTCAGGCCTGTCG.................................................................................. | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................TCTTCTTGGCTGGAGGAAG.................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................AAGAATGATGTCAGGCCTGACGGCA............................................................................... | 25 | 1 | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................AGTGTATTTAACATACAA.................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .......................................ATATTCAGAATGTGAG................................................................................................................................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| ...............................................................................................................................TGATCATTGCAAAGC............................................................................................................ | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| GGTCTCCCTCACTCCAGAAGAATTTGACCAACTCCAGAAATATTCAGAATGTGAGTAAATGCATTGACAGCCCTGGGACGGGATTTCTTCTTGGCTGGAGAAATCATTTCTGGAAAGCATAGCAGTATGATCATTGCAAAGCCACAAAGAATGATGTCAGGCCTGACGGCTTATATTTCCTTATAAACACAGAATTAGTGTATTTAACATAAAAACGCAATTATCTTTAAGCTCAGTAAATACTACGTGC .................................................................................(((((((((......)))))))))..((((.....((...(((((......)))))...)).....)))).......(((((....))))).............................................................................. ................................................................................81.........................................................................................172............................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM532874(GSM532874) G699T. (cervix) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................AAGAATTTGACCAACTGT....................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ........................................................................................................................................CAAAGCCACAAAGAATGTA............................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |