| (1) AGO3.ip | (4) BRAIN | (15) BREAST | (5) CELL-LINE | (1) CERVIX | (1) HEART | (2) LIVER | (1) OTHER | (38) SKIN | (2) UTERUS |
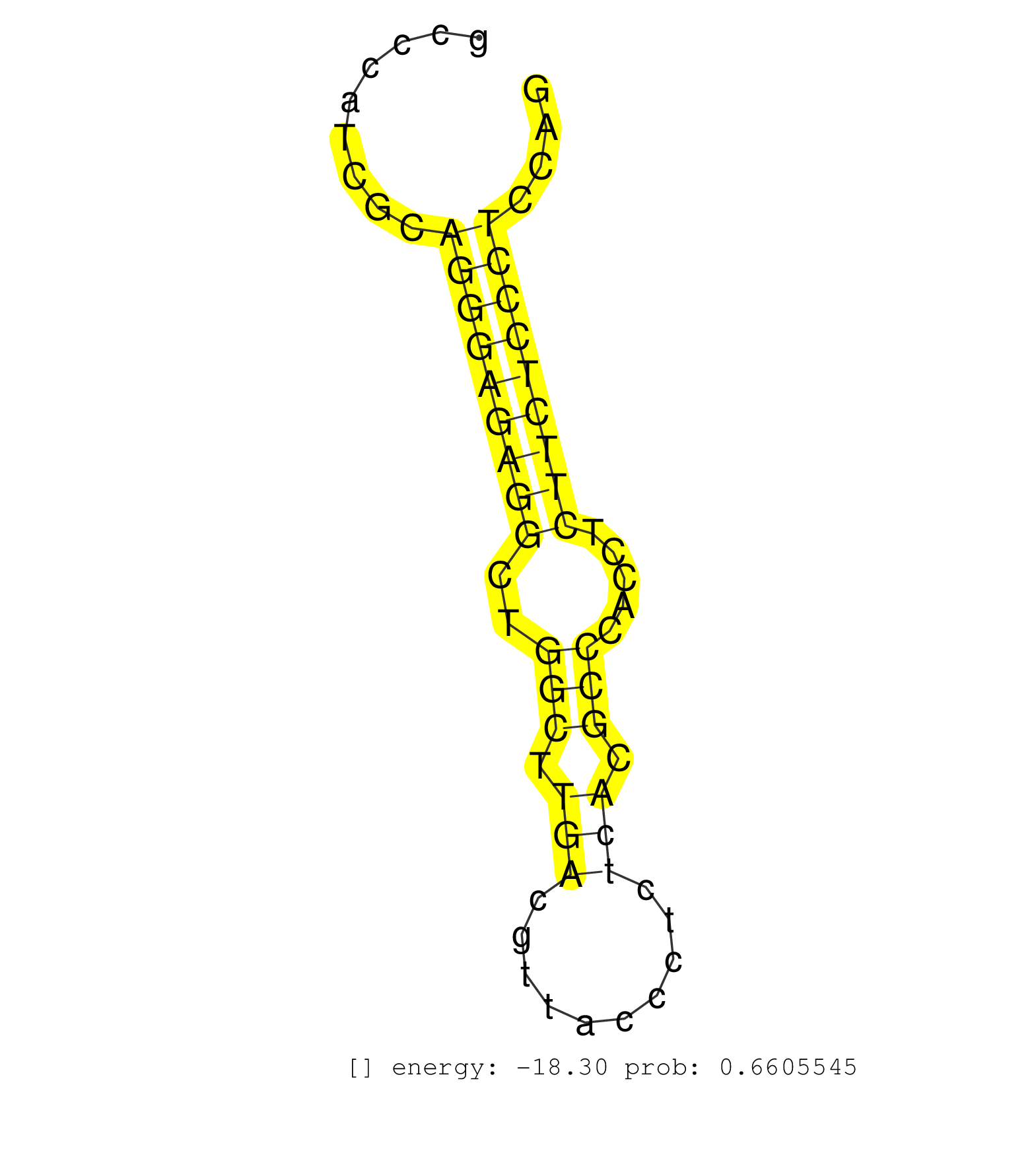
| TGAGAGAGAAAAACTAGGAGTAGAATCCTAGGTGTCTAGCCTTTATGGTCACCAGGAGTTCTCATGGGAGCTGGAGGATTAGGGCAGCAACACAGAGGAATATGGGGCTGGGCTCAGCAGGGAGCCTGCTGGAGCTGTGCCCATCGCAGGGAGAGGCTGGCTTGACGTTACCCTCTCACGCCCACCTCTTCTCCCTCCAGTTGCTCCTGGAATGAAGGTTTATATTGACCCTTTTACCTACGAGGACCCT ...................................................................................................................................................(((((((((..(((.(((..........))).))).....)))))))))...................................................... ..........................................................................................................................................139..........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191557(GSM715667) 57genomic small RNA (size selected RNA from t. (breast) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189782 | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR191469(GSM715579) 117genomic small RNA (size selected RNA from . (breast) | TAX577588(Rovira) total RNA. (breast) | SRR191506(GSM715616) 146genomic small RNA (size selected RNA from . (breast) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR191421(GSM715531) 122genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | SRR040039(GSM532924) G531T. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR191600(GSM715710) 90genomic small RNA (size selected RNA from t. (breast) | SRR038857(GSM458540) D20. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191533(GSM715643) 136genomic small RNA (size selected RNA from . (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191572(GSM715682) 66genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCAGT................................................. | 24 | 1 | 36.00 | 36.00 | - | 2.00 | 2.00 | - | - | 1.00 | - | 2.00 | - | 1.00 | 1.00 | - | 1.00 | 2.00 | 1.00 | 2.00 | - | 2.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTGA..................................................................................... | 22 | 1 | 10.00 | 10.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCAG.................................................. | 23 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCAGA................................................. | 24 | 1 | 5.00 | 7.00 | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCA................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTGAT.................................................................................... | 23 | 1 | 3.00 | 10.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CGCAGGGAGAGGCTGGCTTGA..................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCC.................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTGAAGT.................................................................................. | 25 | 1 | 2.00 | 10.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTG...................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................TAGAATCCTAGGTGTCTAGCCTTT.............................................................................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................CAGGGAGAGGCTGGCTTGA..................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTTA..................................................................................... | 22 | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................CTCATGGGAGCTGGAGGATT.......................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GGCAGCAACACAGAGGAATATGGGGCT............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTGT..................................................................................... | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCGT.................................................. | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................CAGGGAGAGGCTGGCTTGAAAA.................................................................................. | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CACGCCCACCTCTTCTCCCTCCAGA................................................. | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTTAAA................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCAT.................................................. | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCAGAA................................................ | 25 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGGGGCTGGGCTCAGAGGC................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCT...................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCGCAGGGAGAGGCTGGCTTGAA.................................................................................... | 23 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCCGA................................................. | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................CAGGGAGAGGCTGGCTTG...................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CGCAGGGAGAGGCTGGCTTT...................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCATT.................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................ATCGCAGGGAGAGGCTGGCTTGA..................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CACAGAGGAATATGGGGCTG............................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCCCGT................................................. | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ACGCCCACCTCTTCTCCCTCA.................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................CCACCTCTTCTCCCTCCAG.................................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GTTGCTCCTGGAATGA................................... | 16 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGAGAGAGAAAAACTAGGAGTAGAATCCTAGGTGTCTAGCCTTTATGGTCACCAGGAGTTCTCATGGGAGCTGGAGGATTAGGGCAGCAACACAGAGGAATATGGGGCTGGGCTCAGCAGGGAGCCTGCTGGAGCTGTGCCCATCGCAGGGAGAGGCTGGCTTGACGTTACCCTCTCACGCCCACCTCTTCTCCCTCCAGTTGCTCCTGGAATGAAGGTTTATATTGACCCTTTTACCTACGAGGACCCT ...................................................................................................................................................(((((((((..(((.(((..........))).))).....)))))))))...................................................... ..........................................................................................................................................139..........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191557(GSM715667) 57genomic small RNA (size selected RNA from t. (breast) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189782 | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR191469(GSM715579) 117genomic small RNA (size selected RNA from . (breast) | TAX577588(Rovira) total RNA. (breast) | SRR191506(GSM715616) 146genomic small RNA (size selected RNA from . (breast) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR191421(GSM715531) 122genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | SRR040039(GSM532924) G531T. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR191600(GSM715710) 90genomic small RNA (size selected RNA from t. (breast) | SRR038857(GSM458540) D20. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191533(GSM715643) 136genomic small RNA (size selected RNA from . (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191572(GSM715682) 66genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................CACGCCCACCTCTTCTGG........................................................ | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ........................................................................................................................................................................................CCTCTTCTCCCTCCAGTTGTAG............................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................CTCCTGGAATGAAGGTTCT............................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................CCTGCTGGAGCTGTGCGCA........................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ....AGAGAAAAACTAGGAGTTCA.................................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................TGTTGCTGCCCTAAT.............................................................................................................................................................. | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |