| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (2) AGO2.ip | (8) B-CELL | (1) BRAIN | (3) BREAST | (27) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (2) HELA | (4) LIVER | (2) OTHER | (1) RRP40.ip | (7) SKIN | (3) UTERUS | (1) XRN.ip |
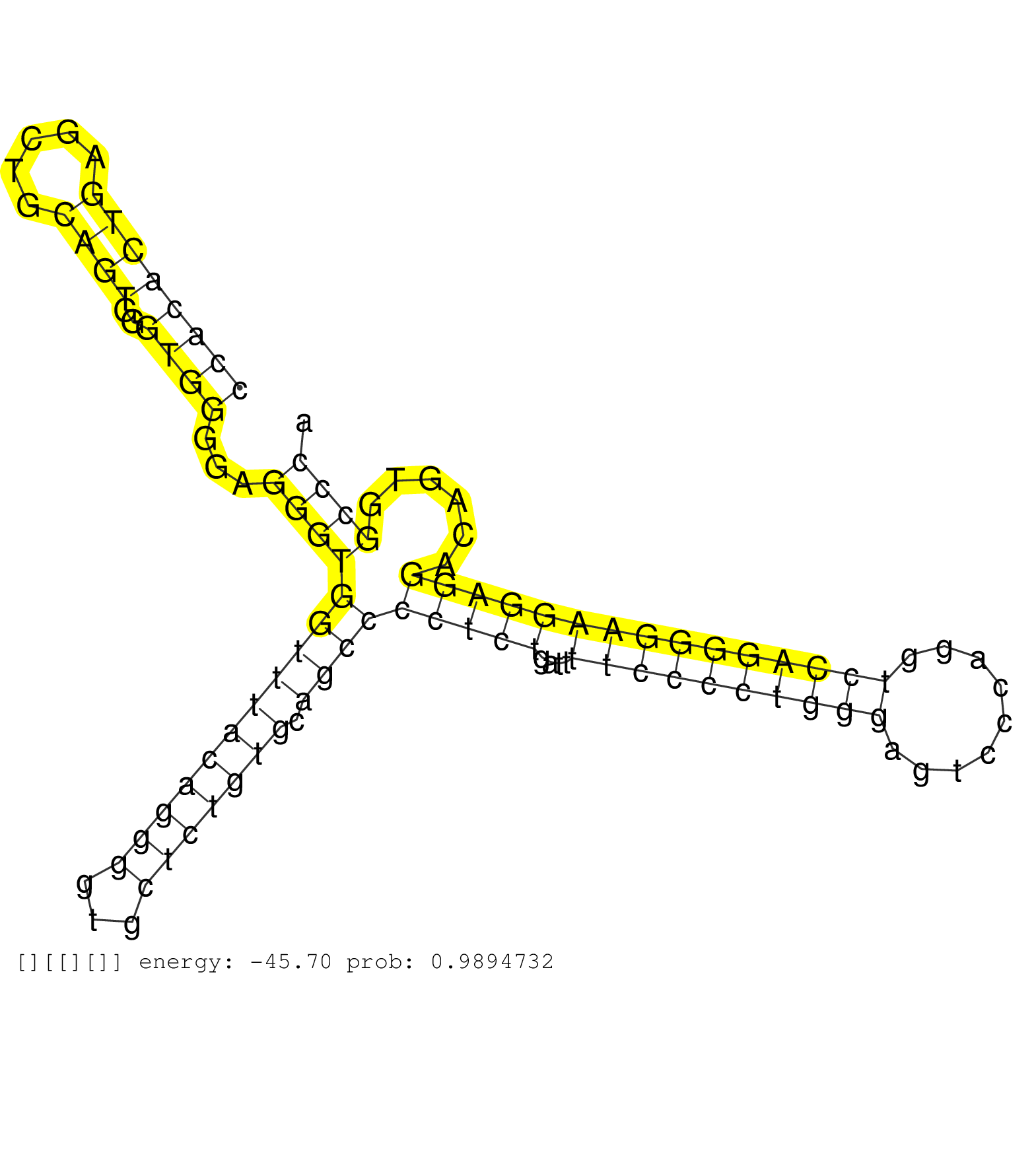
| GATGGGGGACTGGGCCCCTGCCTGGGGGATTGGGTCGTGACCTGTGTGGAGCCCCACACTGAGCTGCAGTGGGTGGGGAGGGTGGTTTACAGGGGTGCTCTGTGCAGCCCCTCTGATTTTCCCCTGGGAGTCCCAGGTCCAGGGGAAGGAGGACAGTGGCCCAGGCCACACAGCTCACTGGGCGGCTCTCACTCCCCCAGGGCTGGCTGCTGGCGGGATGGACACCCTGGAGGAGGTGACTTGGGCCAAT .......................................................................................................................................(((((.(((...((((((.(((.((((((((......))...)))))).))).)).))))))).))))).............................................. .......................................................................................................................................136....................................................................207......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR038856(GSM458539) D11. (cell line) | SRR189784 | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR040025(GSM532910) G613T. (cervix) | SRR038854(GSM458537) MM653. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038859(GSM458542) MM386. (cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR038860(GSM458543) MM426. (cell line) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | TAX577745(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAGT..................................................................................... | 25 | 1 | 7.00 | 2.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGG........................................................................................... | 20 | 2 | 6.50 | 6.50 | - | 2.50 | 2.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - |
| ..........................................................................................................................................CCAGGGGAAGGAGGACAGTGG........................................................................................... | 21 | 1 | 5.00 | 5.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGCCCAAT..................................................................................... | 26 | 5.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCC......................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TCCAGGGGAAGGAGGACAGTGG........................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGG........................................................................................... | 19 | 2 | 3.00 | 3.00 | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 |
| ..........................................................................................................................................CCAGGGGAAGGAGGACAGTGGAAA........................................................................................ | 24 | 1 | 3.00 | 5.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGT............................................................................................. | 18 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - | - |
| .....................................................................................................................................................................................GCGGCTCTCACTCCCCCAGAAC............................................... | 22 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGC.......................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGC.......................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAG...................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................CCCTGGAGGAGGTGACTTGGG..... | 21 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AGGCCACACAGCTCACTGG..................................................................... | 19 | 3 | 1.67 | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGCCCAAA..................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAGA..................................................................................... | 25 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CCAGGGGAAGGAGGACAGTGGCC......................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAGGT.................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................CTGAGCTGCAGTGGGTGGGGAGGGTGG..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAGGAGGACAGTGGCCCAGGT.................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................CCAGGGCTGGCTGCTGGCGGGATGGACACC........................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................ACTCCCCCAGGGCTGGAAG......................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCC........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................GGGTGGGGAGGGTGG..................................................................................................................................................................... | 15 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GGACACCCTGGAGGAGGTGA........... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGCC......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGCCCATC..................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................TCCAGGGGAAGGAGGACAGTGGA.......................................................................................... | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TCCAGGGGAAGGAGGACAGGGA........................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAAGA.................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTG............................................................................................ | 18 | 2 | 1.00 | 1.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| .........................................................................................................................................TCCAGGGGAAGGAGGACAGGGG........................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAGC..................................................................................... | 25 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCAA....................................................................................... | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAT...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CCAGGGGAAGGAGGACAGTGGCCCAAT..................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................GCTGGCGGGATGGACACC........................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AGGAGGACAGTGGCCACCC..................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCTA...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CCAGGGGAAGGAGGACAGTGA........................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCAA...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................ATGGACACCCTGGAGGATG.............. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................TCCAGGGGAAGGAGGACAGTGA........................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................CTGGAGGAGGTGACTCTGA..... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGAAA........................................................................................ | 22 | 2 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGCAA........................................................................................ | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................CACTCCCCCAGGGCTTTAG.......................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................TCCAGGGGAAGGAGGACAGT............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGCCCA....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGCCCAGT..................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................AGCTGCAGTGGGTGGTGG........................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGGGAAGGAGGACAGT............................................................................................. | 17 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGTGGAA......................................................................................... | 22 | 2 | 0.50 | 6.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGA........................................................................................... | 19 | 2 | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ............................................................................................................................................AGGGGAAGGAGGACAGTGGAA......................................................................................... | 21 | 2 | 0.50 | 3.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAGCGG........................................................................................... | 20 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGGGGAAGGAGGACAG.............................................................................................. | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - |
| GATGGGGGACTGGGCCCCTGCCTGGGGGATTGGGTCGTGACCTGTGTGGAGCCCCACACTGAGCTGCAGTGGGTGGGGAGGGTGGTTTACAGGGGTGCTCTGTGCAGCCCCTCTGATTTTCCCCTGGGAGTCCCAGGTCCAGGGGAAGGAGGACAGTGGCCCAGGCCACACAGCTCACTGGGCGGCTCTCACTCCCCCAGGGCTGGCTGCTGGCGGGATGGACACCCTGGAGGAGGTGACTTGGGCCAAT .......................................................................................................................................(((((.(((...((((((.(((.((((((((......))...)))))).))).)).))))))).))))).............................................. .......................................................................................................................................136....................................................................207......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR038856(GSM458539) D11. (cell line) | SRR189784 | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR040025(GSM532910) G613T. (cervix) | SRR038854(GSM458537) MM653. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038859(GSM458542) MM386. (cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR038860(GSM458543) MM426. (cell line) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | TAX577745(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................CCCAGGTCCAGGGGAACGC.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................CCCCTGTAAACCACCCTCCCCACCCAC........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |