| (1) AGO2.ip | (1) B-CELL | (2) BREAST | (7) CELL-LINE | (1) HEART | (2) HELA | (3) LIVER | (2) OTHER | (1) UTERUS |
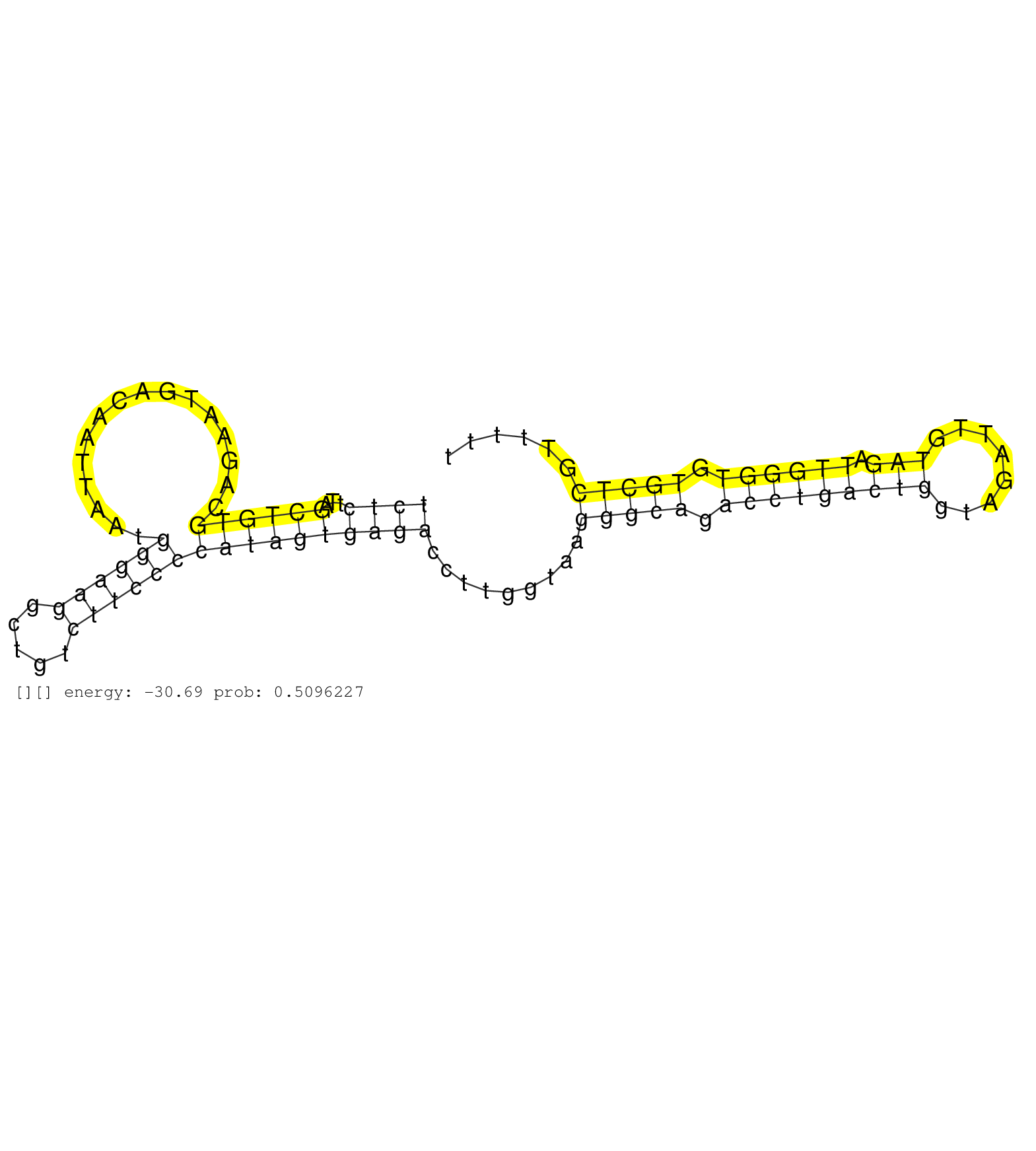
| TCCTGTCCCTTCCTGGGGTGTGTGTCGGTTGCCCTGAGCGACCGGCCATGGGACTCTGTCGTGATAACCAAGCTTCAGGGTGTGGGAAGAGGACAGTCAGTGCTTCCTTGGGGCATCACTCGGTAACATCATGGGCATAAACAAAAGTACTCAGTCTTCAAGGTCATAAAGTAACCAGAGTGTATTCCTTTTTTTTTCAGATCTCTTACCTCAGCTAGAAGCTCCGAGTTCTCTTACTCCCAGCAGTGAA ...............................................................................................................((.((((((((((.((....((((.((........)).))))..((....)).......)).))).))))).)).)).............................................................. ...............................................................................................................112............................................................................................207......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR189782 | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189784 | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR343334 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................ATTCCTTTTTTTTTCAAATG............................................... | 20 | 20.00 | 0.00 | 7.00 | 2.00 | 9.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TATTCCTTTTTTTTTCAAATG............................................... | 21 | 20.00 | 0.00 | 7.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................CATCATGGGCATAAACAAAAGTACTCAGTCTTCAAGGTCATAAAGTAACCAGAGTGTATTCCTTTTTTTT...................................................... | 70 | 1 | 6.00 | 6.00 | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TATTCCTTTTTTTTTAATG................................................. | 19 | 4.00 | 0.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................ACATCATGGGCATAAACAAAAGTACTCAGTCTTCAAGGTCATAAAGTAACCAGAGTGTATTCCTTTTTTTTTCAG.................................................. | 75 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AGGACAGTCAGTGCTTCCTTGGGGCACC..................................................................................................................................... | 28 | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TATTCCTTTTTTTTTCAAACG............................................... | 21 | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................ACCAAGCTTCAGGGTGTGGGAAGAGGACAGCC........................................................................................................................................................ | 32 | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................ATTCCTTTTTTTTTCAAACG............................................... | 20 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................CTTACCTCAGCTAGAAGCTCCGAGTTCTCT................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TATTCCTTTTTTTTTCAATG................................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................ATTCCTTTTTTTTTCAGACGA.............................................. | 21 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................ATTCCTTTTTTTTTCAATAG............................................... | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................CACTCGGTAACATCATGGGCATAAA............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................AGCTTCAGGGTGTGGGAAGAGG.............................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................GAGGACAGTCAGTGCAAC................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .......................................................................................................................................................................................ATTCCTTTTTTTTTCAATTC............................................... | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................TCCTTTTTTTTTCAGACGG.............................................. | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................TCTCTTACTCCCAGCTTCG.. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................TTCTCTTACTCCCAGCAG.... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AGAAGCTCCGAGTTCTCTT............... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGGGGTGTGTGTCGGAGGG.......................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......CCCTTCCTGGGGTGTGGAGA................................................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .....................................................................................................................................................................................................................GCTAGAAGCTCCGAGTCTAG................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................TTCCTTTTTTTTTCAGAATAT............................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................AGCTTCAGGGTGTGGGAAGAGGACAGCCA....................................................................................................................................................... | 29 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TATTCCTTTTTTTTTCAATAG............................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TATTCCTTTTTTTTTCAAAGG............................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................AAGCTTCAGGGTGTGGGGTAG................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........CCTGGGGTGTGTGTCGGT............................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TAGAAGCTCCGAGTTCTCT................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................................ATTCCTTTTTTTTTCAACCG............................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TATTCCTTTTTTTTTCAG.................................................. | 18 | 7 | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................GTCAGTGCTTCCTTG............................................................................................................................................ | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| TCCTGTCCCTTCCTGGGGTGTGTGTCGGTTGCCCTGAGCGACCGGCCATGGGACTCTGTCGTGATAACCAAGCTTCAGGGTGTGGGAAGAGGACAGTCAGTGCTTCCTTGGGGCATCACTCGGTAACATCATGGGCATAAACAAAAGTACTCAGTCTTCAAGGTCATAAAGTAACCAGAGTGTATTCCTTTTTTTTTCAGATCTCTTACCTCAGCTAGAAGCTCCGAGTTCTCTTACTCCCAGCAGTGAA ...............................................................................................................((.((((((((((.((....((((.((........)).))))..((....)).......)).))).))))).)).)).............................................................. ...............................................................................................................112............................................................................................207......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR189782 | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189784 | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR343334 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................GAGTTCTCTTACTCCCATG...... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .................................................................................................................................CATGGGCATAAACAAGAA....................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAAGACTGAGTACTTTTGT.......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................ACTCAGTCTTCAAGGTG..................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................TCAGTCTTCAAGGTCAGTG................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |