| (1) BRAIN | (3) BREAST | (12) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (1) HEART | (2) HELA | (1) LIVER | (2) OTHER | (7) SKIN | (1) TESTES | (1) XRN.ip |
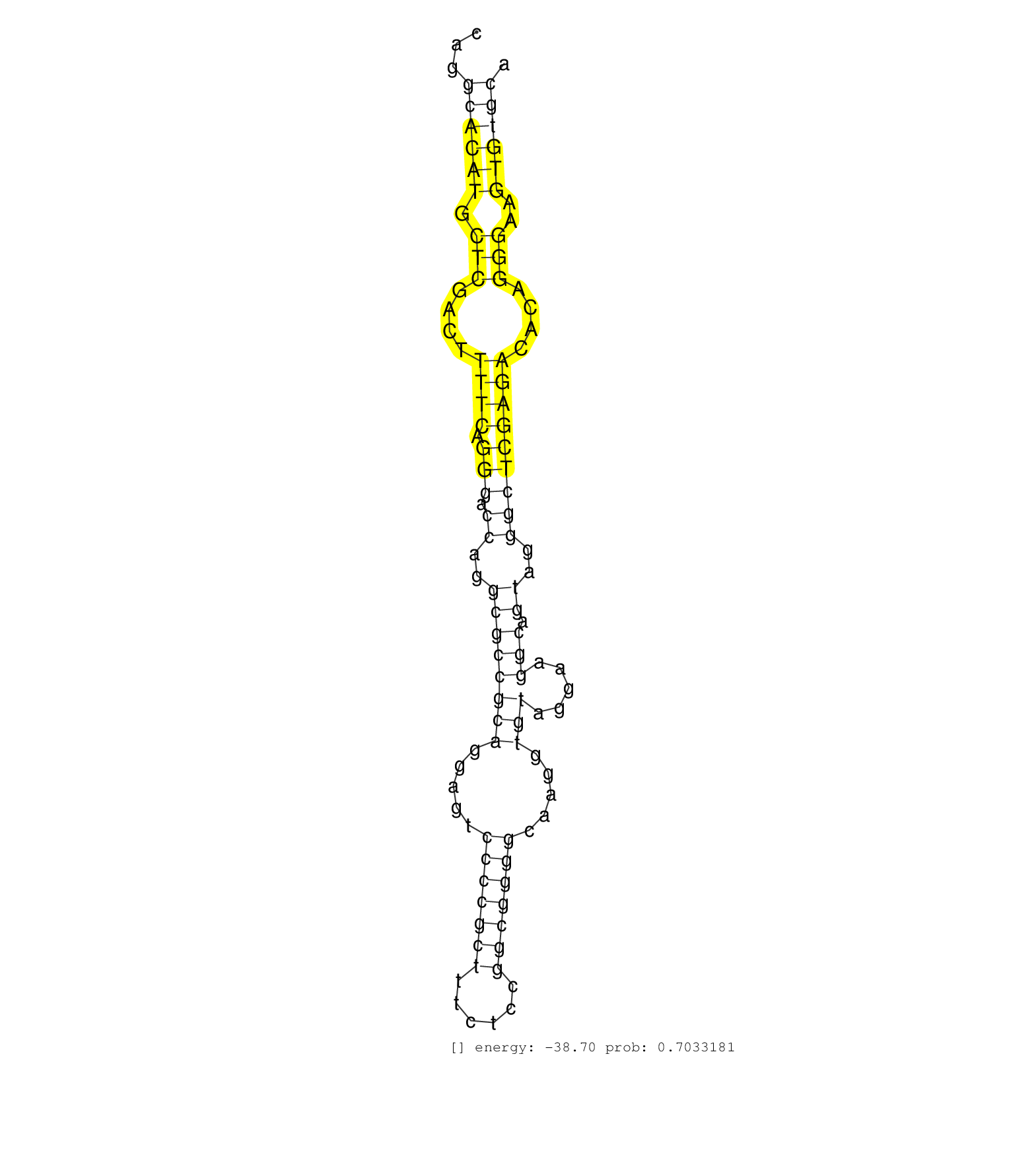
| AATGAGTGGTCAGGTAGTCTTAAAGAGCCTCATGTTAAATAGACACAAATTTGCATAGTTGAATTCTTTAATAGACTTAATTTAAGATTTTGTGGGGTTTTTTTGAGAAATTAATGGCTTAATAAAATGGATAATTACAACATGGGCTTAAAACATAGAACTATTTACAAATCTTATTTTCCTTAGCTAAAAACCTTTAGGTATCTTCTTATTTTCTTGAAATTTGTACAAGCTGTCATTCCAACAATAG ................................................................................................................................................((((........(((............)))..........)))).............................................................. .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189785 | SRR037931(GSM510469) 293GFP. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | SRR029129(GSM416758) SW480. (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR553576(SRX182782) source: Testis. (testes) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM532874(GSM532874) G699T. (cervix) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................TTTCCTTAGCTAAAAACCTTTAGA................................................. | 24 | 1 | 17.00 | 5.00 | 11.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ........................................................................................................................................ACAACATGGGCTTAAAACATAGAACTATTTACAAATCTTATTTTCCTTAGCTAAAAACCTTTAG.................................................. | 64 | 1 | 7.00 | 7.00 | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CAACATGGGCTTAAAACATAGAACTATTTACAAATCTTATTTTCCTTAGCTAAAAACCTTTAG.................................................. | 63 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTCCTTAGCTAAAAACCTTTAG.................................................. | 23 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AACTATTTACAAATCTTATTTTCCTTAG................................................................ | 28 | 1 | 5.00 | 5.00 | - | - | - | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGGCTTAAAACATAGAACTA....................................................................................... | 21 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTCCTTAGCTAAAAACC....................................................... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................ATTTTCCTTAGCTAAAAACCTTTAG.................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AGTGGTCAGGTAGTCTTA.................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TTTTCCTTAGCTAAAAACCTTTAGA................................................. | 25 | 2.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................TTTGTACAAGCTGTCATTCCA....... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................TTTTGTGGGGTTTTTTCGT................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................TACAAGCTGTCATTCCAACAATAG | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................AAAAACCTTTAGGTATTT............................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ...............................................................................................................................................................................................................................TTGTACAAGCTGTCATTCCAAC..... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCTTATTTTCTTGAAATTG......................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .................................................................................................................................................................................TTTCCTTAGCTAAAAACCTTTAGAA................................................ | 25 | 1 | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................AGATTTTGTGGGGTTTTTCCC................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................GCTTAAAACATAGAACTCTG..................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .......................................................................................TTTTGTGGGGTTTTTGCAT................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................AAATTTGTACAAGCTGTTATT.......... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ...............................................................................ATTTAAGATTTTGTGGTAAG....................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................ATCTTATTTTCCTTAGCTAAAAACCTTTAG.................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ATCTTCTTATTTTCTTGAAAT........................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ........................................................................................................................................................................................................GTATCTTCTTATTTTCTT................................ | 18 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AATGAGTGGTCAGGTAGTCTTAAAGAGCCTCATGTTAAATAGACACAAATTTGCATAGTTGAATTCTTTAATAGACTTAATTTAAGATTTTGTGGGGTTTTTTTGAGAAATTAATGGCTTAATAAAATGGATAATTACAACATGGGCTTAAAACATAGAACTATTTACAAATCTTATTTTCCTTAGCTAAAAACCTTTAGGTATCTTCTTATTTTCTTGAAATTTGTACAAGCTGTCATTCCAACAATAG ................................................................................................................................................((((........(((............)))..........)))).............................................................. .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189785 | SRR037931(GSM510469) 293GFP. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | SRR029129(GSM416758) SW480. (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR553576(SRX182782) source: Testis. (testes) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM532874(GSM532874) G699T. (cervix) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................CACAAAATCTTAAATTA............................................................................................................................................................ | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |