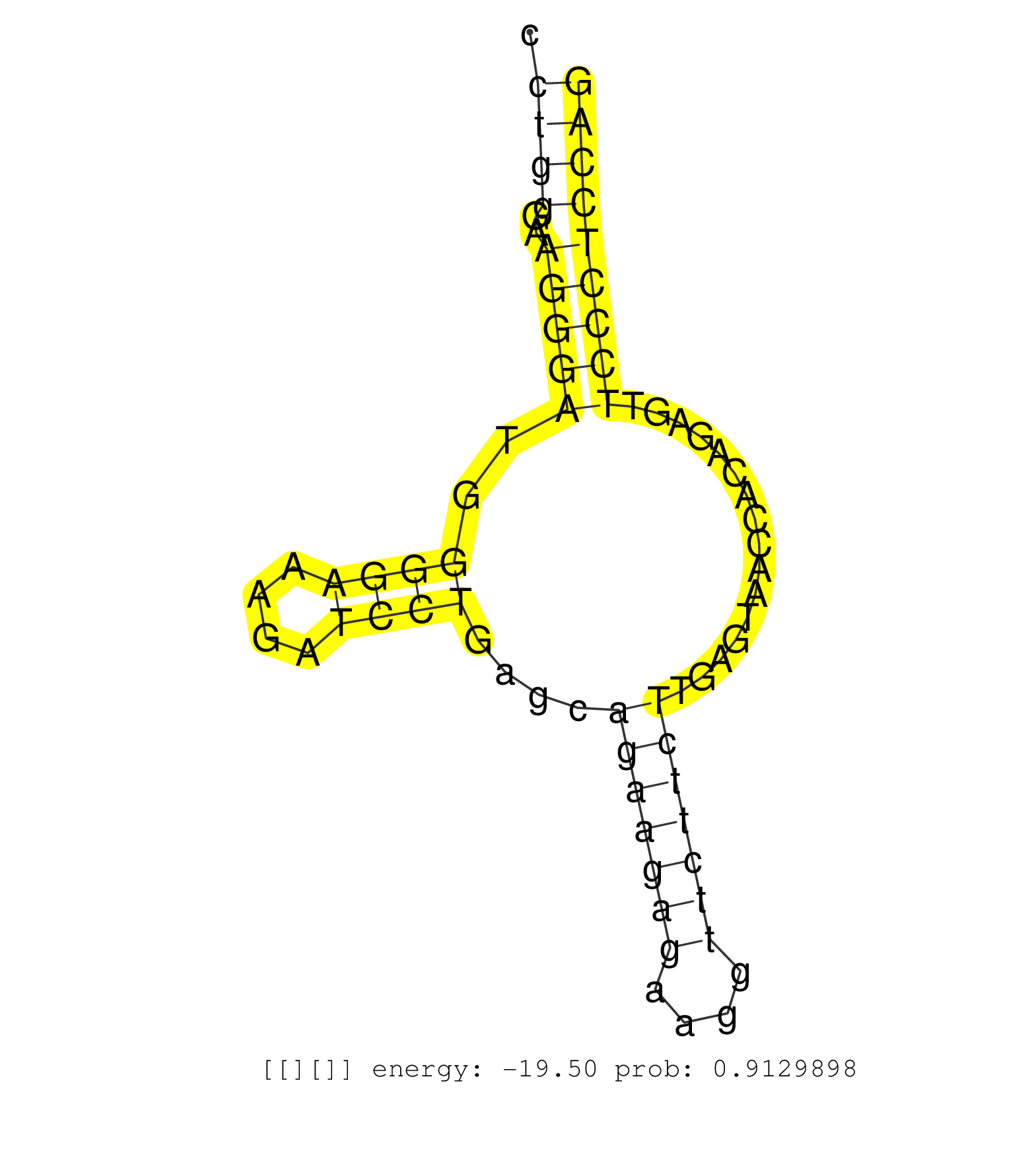
| (1) AGO2.ip | (1) B-CELL | (6) BRAIN | (28) BREAST | (15) CELL-LINE | (16) CERVIX | (1) FIBROBLAST | (5) HEART | (3) HELA | (4) LIVER | (1) OTHER | (8) SKIN | (1) XRN.ip |
| TGACCTGCCCTGGCGTCCGGCTGAGGACGGCCATAGTCATGCCTGTAGGAGTGAGTGAGATCTTGGCCTCCATAGCTCCAGCACTCTGCAGCCTGGCTGTGCCCTGGGTAGAACCTAGAGCTAGTTGGTCAAGCCCCTAAAACCCAGGCTGTATCCAAAGAGTATGCAGACCTTGATGCCCCTTGGGACCCCCTCAATCCTCCTCTGCAGTCTGCTCTGGGTAGGGAACCAGTATGATGCTGAGGCCTTGAAGTCCCTGAGCCCCTACCCACAAATTCTGGCCTGTGATAGGGATCTAAGGTGGCCTACTTCTTACCCTTGTGACCTGGTTCCTCATGTGGCCTTTGCCCTGCCCTTCCTCCCCTGCCAGAGTGTGAGTGCCATGGGCACACCCACAGCTGCCACTTCGACATGGCCGTA .....................................................................................................................................................................................((((((((....((((....((((.(((.((....((((........))))...))))).))))..)))).))))).)))............................................................................................................................................................... .............................................................................................................................................................................174....................................................................................................277............................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR040016(GSM532901) G645N. (cervix) | SRR037938(GSM510476) 293Red. (cell line) | SRR040024(GSM532909) G613N. (cervix) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR189782 | TAX577744(Rovira) total RNA. (breast) | SRR040018(GSM532903) G701N. (cervix) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | TAX577590(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040006(GSM532891) G601N. (cervix) | SRR040012(GSM532897) G648N. (cervix) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | TAX577743(Rovira) total RNA. (breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR029124(GSM416753) HeLa. (hela) | TAX577580(Rovira) total RNA. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189785 | TAX577742(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR191461(GSM715571) 35genomic small RNA (size selected RNA from t. (breast) | SRR040032(GSM532917) G603N. (cervix) | SRR191453(GSM715563) 179genomic small RNA (size selected RNA from . (breast) | TAX577745(Rovira) total RNA. (breast) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR040030(GSM532915) G013N. (cervix) | SRR191485(GSM715595) 123genomic small RNA (size selected RNA from . (breast) | SRR191628(GSM715738) 52genomic small RNA (size selected RNA from t. (breast) | SRR191397(GSM715507) 30genomic small RNA (size selected RNA from t. (breast) | SRR040010(GSM532895) G529N. (cervix) | SRR191441(GSM715551) 106genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM359188(GSM359188) HepG2_2pM_2. (cell line) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040011(GSM532896) G529T. (cervix) | GSM532875(GSM532875) G547N. (cervix) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM532874(GSM532874) G699T. (cervix) | SRR191500(GSM715610) 7genomic small RNA (size selected RNA from to. (breast) | GSM359207(GSM359207) HepG2_tot_up. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR191481(GSM715591) 43genomic small RNA (size selected RNA from t. (breast) | SRR040031(GSM532916) G013T. (cervix) | SRR040008(GSM532893) G727N. (cervix) | TAX577739(Rovira) total RNA. (breast) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR040022(GSM532907) G575N. (cervix) | SRR191548(GSM715658) 101genomic small RNA (size selected RNA from . (breast) | SRR040033(GSM532918) G603T. (cervix) | GSM359205(GSM359205) HepG2_nucl_up. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR191530(GSM715640) 132genomic small RNA (size selected RNA from . (breast) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029132(GSM416761) MB-MDA231. (cell line) | TAX577589(Rovira) total RNA. (breast) | SRR191431(GSM715541) 168genomic small RNA (size selected RNA from . (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | TAX577738(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR040020(GSM532905) G699N_2. (cervix) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191472(GSM715582) 120genomic small RNA (size selected RNA from . (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR191446(GSM715556) 140genomic small RNA (size selected RNA from . (breast) | SRR191532(GSM715642) 135genomic small RNA (size selected RNA from . (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTTGT................................................................................................................................................... | 20 | 55.00 | 0.00 | 7.00 | - | 3.00 | - | 4.00 | - | - | - | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | 2.00 | 2.00 | - | 3.00 | 2.00 | - | - | - | - | - | 2.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTTG.................................................................................................................................................... | 19 | 12.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................CCCTGAGCCCCTACCTTGT................................................................................................................................................... | 19 | 7.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................TATGATGCTGAGGCCTTGAAGTCCCTGAGC.............................................................................................................................................................. | 30 | 1 | 6.00 | 6.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TAGTCATGCCTGTAGGA.................................................................................................................................................................................................................................................................................................................................................................................. | 17 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCCTGT................................................................................................................................................... | 20 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................CAGTATGATGCTGAGGCCTTGAAGTCCCTGAGC.............................................................................................................................................................. | 33 | 1 | 4.00 | 4.00 | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................CCAGTATGATGCTGAGGCCTTGAAGTCCCTGA................................................................................................................................................................ | 32 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................CCCTGAGCCCCTACCTTTT................................................................................................................................................... | 19 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTTGA................................................................................................................................................... | 20 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCCTGTG.................................................................................................................................................. | 21 | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................TATGATGCTGAGGCCTTGAAGTCCCTGA................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CCCCTTGGGACCCCCTCAATCC............................................................................................................................................................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CAGTATGATGCTGAGGCCTTGAAGT...................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CTCAATCCTCCTCTGCAGTCT............................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CAGTATGATGCTGAGGCCTTGAAGTCCCTGAG............................................................................................................................................................... | 32 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................AATTCTGGCCTGTGATAGGGATCTAAGG....................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGCGTCCGGCTGAGGACGGCCA................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GAGGACGGCCATAGTCATGC.......................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................GGGACCCCCTCAATCCTCC......................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................CATGGGCACACCCACAGCTGCC................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................CTGGGTAGAACCTAGAGCTAGTTGGTC.................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TCTGCTCTGGGTAGGGGA................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................................................................................................GCTGCCACTTCGACAC....... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................AGTCATGCCTGTAGGGGC................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTTA.................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTT..................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................GTATGATGCTGAGGCCTTGAAGT...................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................CGGCTGAGGACGGCCAT.................................................................................................................................................................................................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GACCTGCCCTGGCGTC................................................................................................................................................................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTCGC................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................................................................................................................................................................AGAGTGTGAGTGCCATACAG................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................GTATGATGCTGAGGCCTTGAAGTCCCTGA................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................ACCAGTATGATGCTGAGGAC............................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| .....................TGAGGACGGCCATAG................................................................................................................................................................................................................................................................................................................................................................................................ | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................AGGACGGCCATAGTCATG........................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTTCT................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCCCATG.................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................CAGTATGATGCTGAGGCCTTGAAGTCCCTGA................................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCT...................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................CAGTATGATGCTGAGGCCTTGA......................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................TCCTCTGCAGTCTGCTGATT........................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................CCAGTATGATGCTGAGGCCTTGAAGTCCCTGCGC.............................................................................................................................................................. | 34 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................GGTAGGGAACCAGTACCGT...................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CCTGGCGTCCGGCTGAG........................................................................................................................................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................ACGGCCATAGTCATGCCTGTAGG................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GGACGGCCATAGTCATGC.......................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................TATGATGCTGAGGCCTTGAAGTCCCTGAG............................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCCTCTG.................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................................................................................................................................................CCTTCCTCCCCTGCCAGAGG............................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............GTCCGGCTGAGGACGGCCATAGTCATGC.......................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACTTTT.................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................................................................................................................................................................AGTGTGAGTGCCATGGGCA............................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GCCTCCATAGCTCCATGG................................................................................................................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTTTT................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................TCCCTGAGCCCCTACCTCGT................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................CCAGTATGATGCTGAGGC.............................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GCAGCCTGGCTGTGCCCTGGGTAG..................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CCCCTTGGGACCCCCTCAA............................................................................................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ................................................................................................................................................................AGTATGCAGACCTTGATG.................................................................................................................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GTCCGGCTGAGGACG....................................................................................................................................................................................................................................................................................................................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................................TGCCATGGGCACACC........................... | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGACCTGCCCTGGCGTCCGGCTGAGGACGGCCATAGTCATGCCTGTAGGAGTGAGTGAGATCTTGGCCTCCATAGCTCCAGCACTCTGCAGCCTGGCTGTGCCCTGGGTAGAACCTAGAGCTAGTTGGTCAAGCCCCTAAAACCCAGGCTGTATCCAAAGAGTATGCAGACCTTGATGCCCCTTGGGACCCCCTCAATCCTCCTCTGCAGTCTGCTCTGGGTAGGGAACCAGTATGATGCTGAGGCCTTGAAGTCCCTGAGCCCCTACCCACAAATTCTGGCCTGTGATAGGGATCTAAGGTGGCCTACTTCTTACCCTTGTGACCTGGTTCCTCATGTGGCCTTTGCCCTGCCCTTCCTCCCCTGCCAGAGTGTGAGTGCCATGGGCACACCCACAGCTGCCACTTCGACATGGCCGTA .....................................................................................................................................................................................((((((((....((((....((((.(((.((....((((........))))...))))).))))..)))).))))).)))............................................................................................................................................................... .............................................................................................................................................................................174....................................................................................................277............................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR040016(GSM532901) G645N. (cervix) | SRR037938(GSM510476) 293Red. (cell line) | SRR040024(GSM532909) G613N. (cervix) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR189782 | TAX577744(Rovira) total RNA. (breast) | SRR040018(GSM532903) G701N. (cervix) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | TAX577590(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040006(GSM532891) G601N. (cervix) | SRR040012(GSM532897) G648N. (cervix) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | TAX577743(Rovira) total RNA. (breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR029124(GSM416753) HeLa. (hela) | TAX577580(Rovira) total RNA. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189785 | TAX577742(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR191461(GSM715571) 35genomic small RNA (size selected RNA from t. (breast) | SRR040032(GSM532917) G603N. (cervix) | SRR191453(GSM715563) 179genomic small RNA (size selected RNA from . (breast) | TAX577745(Rovira) total RNA. (breast) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR040030(GSM532915) G013N. (cervix) | SRR191485(GSM715595) 123genomic small RNA (size selected RNA from . (breast) | SRR191628(GSM715738) 52genomic small RNA (size selected RNA from t. (breast) | SRR191397(GSM715507) 30genomic small RNA (size selected RNA from t. (breast) | SRR040010(GSM532895) G529N. (cervix) | SRR191441(GSM715551) 106genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM359188(GSM359188) HepG2_2pM_2. (cell line) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040011(GSM532896) G529T. (cervix) | GSM532875(GSM532875) G547N. (cervix) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM532874(GSM532874) G699T. (cervix) | SRR191500(GSM715610) 7genomic small RNA (size selected RNA from to. (breast) | GSM359207(GSM359207) HepG2_tot_up. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR191481(GSM715591) 43genomic small RNA (size selected RNA from t. (breast) | SRR040031(GSM532916) G013T. (cervix) | SRR040008(GSM532893) G727N. (cervix) | TAX577739(Rovira) total RNA. (breast) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR040022(GSM532907) G575N. (cervix) | SRR191548(GSM715658) 101genomic small RNA (size selected RNA from . (breast) | SRR040033(GSM532918) G603T. (cervix) | GSM359205(GSM359205) HepG2_nucl_up. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR191530(GSM715640) 132genomic small RNA (size selected RNA from . (breast) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029132(GSM416761) MB-MDA231. (cell line) | TAX577589(Rovira) total RNA. (breast) | SRR191431(GSM715541) 168genomic small RNA (size selected RNA from . (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | TAX577738(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR040020(GSM532905) G699N_2. (cervix) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191472(GSM715582) 120genomic small RNA (size selected RNA from . (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR191446(GSM715556) 140genomic small RNA (size selected RNA from . (breast) | SRR191532(GSM715642) 135genomic small RNA (size selected RNA from . (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................GGGACCCCCTCAATCCTCTGG....................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................CTTCGACATGGCCGTCG | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ...............................................................................................................................................................................................................................................................................................................TCACAAGGGTAAGAAGTAGGC................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................ATACTCTTTGGATACAG............................................................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......GCCGGACGCCAGGGC............................................................................................................................................................................................................................................................................................................................................................................................................... | 15 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |