| (6) B-CELL | (11) BRAIN | (6) BREAST | (13) CELL-LINE | (1) CERVIX | (1) HEART | (1) HELA | (6) LIVER | (1) OTHER | (1) RRP40.ip | (8) SKIN | (3) UTERUS | (1) XRN.ip |
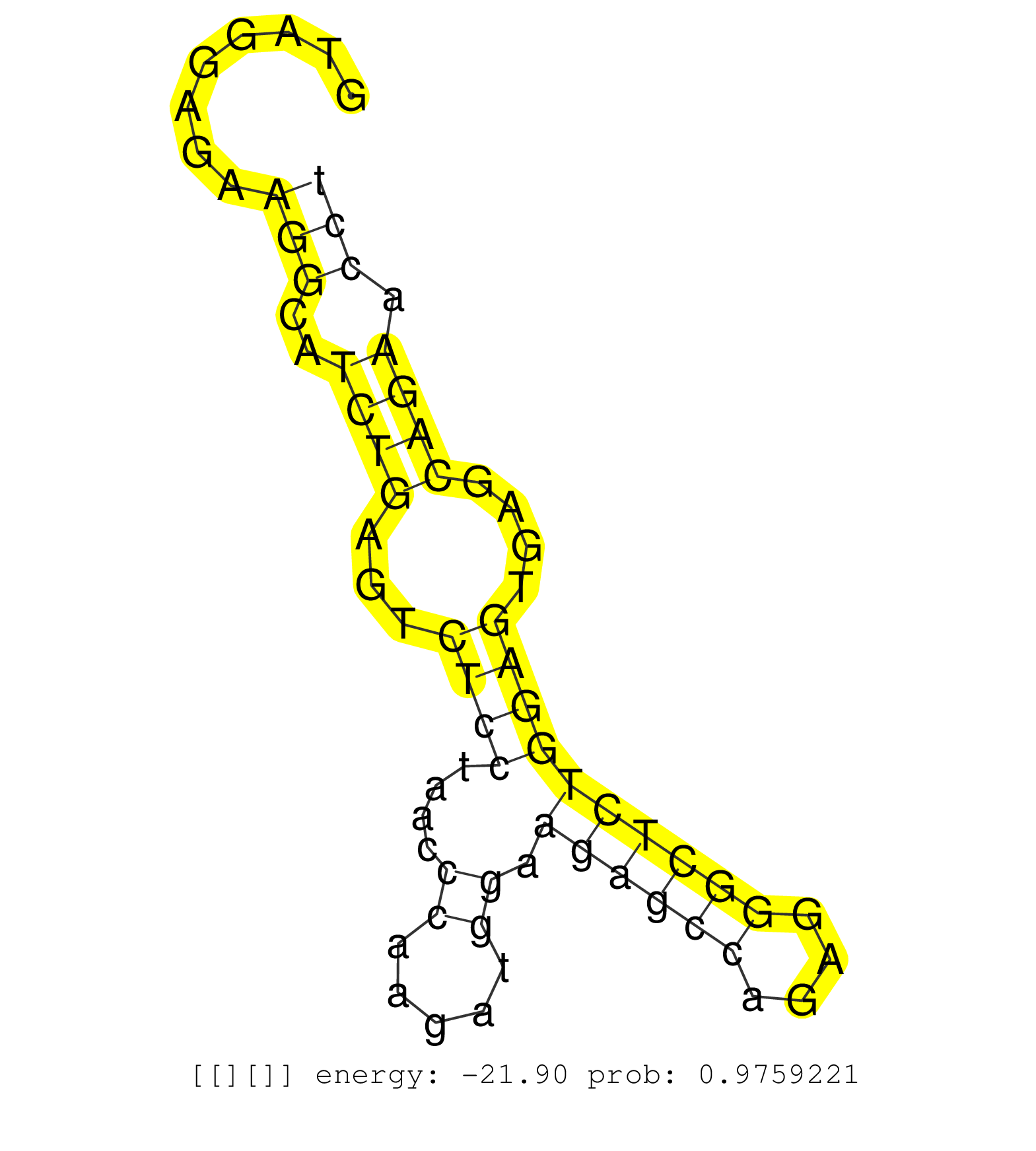
| AGAGTCTACCCCATCGCCAGTCGTCTTCTGCCACAATGACATCCAGGAAGGTAGGAGAAGGCATCTGAGTCTCCTAACCCAAGATGGAAGAGCCAGAGGGCTCTGGAGTGAGCAGAACCTCACCCCATTCCCCCAGGGAACATCTTGCTGCTCTCAGAGCCAGAAAATGCTGACAGCCTCATGCTG ...................................................(((((((.((...))...))))))).....(((((..((((((....))))))((.(((((......))))).))..((((...)))).)))))......................................... ..................................................51............................................................................................145....................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR189784 | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577743(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | GSM532876(GSM532876) G547T. (cervix) | TAX577588(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR189785 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | TAX577744(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGAGAAGGCATCTGAGTCT.................................................................................................................. | 22 | 1 | 21.00 | 21.00 | 5.00 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAGT.................................................................................................................... | 20 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAAA.................................................................................................................... | 20 | 1 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGA...................................................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................CATCCAGGAAGGTAGCAGC................................................................................................................................ | 19 | 3.00 | 0.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTAGGAGAAGGCATCTGAA..................................................................................................................... | 19 | 1 | 2.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAGTCA.................................................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAGTCTC................................................................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAGTC................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................ACAATGACATCCAGGAAG........................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................CATCCAGGAAGGTAGGAGAA............................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTTCTGCCACAATGACATCCAGG........................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAAAT................................................................................................................... | 21 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACATCTTGCTGCTCTCAGAG........................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................ATGACATCCAGGAAGGTAGGAGA................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AGGGAACATCTTGCTTGTC................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................AAGGCATCTGAGTCTCCT............................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................CACCCCATTCCCCCAGGGAAC............................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATCCAGGAAGGTAGGAGAAGGCATCTGAG..................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCTACCCCATCGCCAGTCGTCTTCTGC........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................CAGGAAGGTAGGAGAAGGC............................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAG..................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAGGCTC................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................CAGGAAGGTAGGAGAAGGCATC......................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TGACATCCAGGAAGGTAGGAG................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CAATGACATCCAGGAAGG....................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGCAAT................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................GAAGGTAGGAGAAGGCATCTGCGTC................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................AGGCATCTGAGTCTCCTA.............................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CAGAGGGCTCTGGAGTGAGCAGAAC.................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AGAGGGCTCTGGAGTCACA......................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTAGGAGAAGGCATCTGAGAAAA................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................CATCCAGGAAGGTAGC................................................................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AGAGGGCTCTGGAGTCGCA......................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AGAGGGCTCTGGAGTAATA......................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTAGGAGAAGGCATCTGATTCT.................................................................................................................. | 22 | 1 | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................CATCCAGGAAGGTAGGAGA................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................CCAGGAAGGTAGGAGAAGGC............................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................GAGGGCTCTGGAGTGAGCAGT...................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTAGGAGAAGGCATCTGATT.................................................................................................................... | 20 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAGAAGGCATCTGAAAAA.................................................................................................................. | 22 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................GACATCCAGGAAGGTAGGAGAAGGCATC......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TCCAGGAAGGTAGGAGAAGGCA........................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................AGGAAGGTAGGAGAAGGC............................................................................................................................ | 18 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CTCTGGAGTGAGCAGAAC.................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ..........................TCTGCCACAATGACAT................................................................................................................................................ | 16 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| AGAGTCTACCCCATCGCCAGTCGTCTTCTGCCACAATGACATCCAGGAAGGTAGGAGAAGGCATCTGAGTCTCCTAACCCAAGATGGAAGAGCCAGAGGGCTCTGGAGTGAGCAGAACCTCACCCCATTCCCCCAGGGAACATCTTGCTGCTCTCAGAGCCAGAAAATGCTGACAGCCTCATGCTG ...................................................(((((((.((...))...))))))).....(((((..((((((....))))))((.(((((......))))).))..((((...)))).)))))......................................... ..................................................51............................................................................................145....................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR189784 | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577743(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | GSM532876(GSM532876) G547T. (cervix) | TAX577588(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR189785 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | TAX577744(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........CCATCGCCAGTCGTCTCA.............................................................................................................................................................. | 18 | 6.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................AGCAGAACCTCACCCCTGT......................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............TCGCCAGTCGTCTTCAG............................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CCCCATCGCCAGTCGGG................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................TCTGGCTCTTCCATCTTG......................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TCACCCCATTCCCCCAACAC............................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................CCATTCCCCCAGGGATGT............................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................TTCTGCCACAATGACCCG............................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............CAGAAGACGACTGGC............................................................................................................................................................ | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |