| (1) AGO2.ip | (3) B-CELL | (1) BRAIN | (5) BREAST | (8) CELL-LINE | (6) CERVIX | (2) HEART | (1) HELA | (1) KIDNEY | (6) LIVER | (1) OTHER | (15) SKIN | (1) XRN.ip |
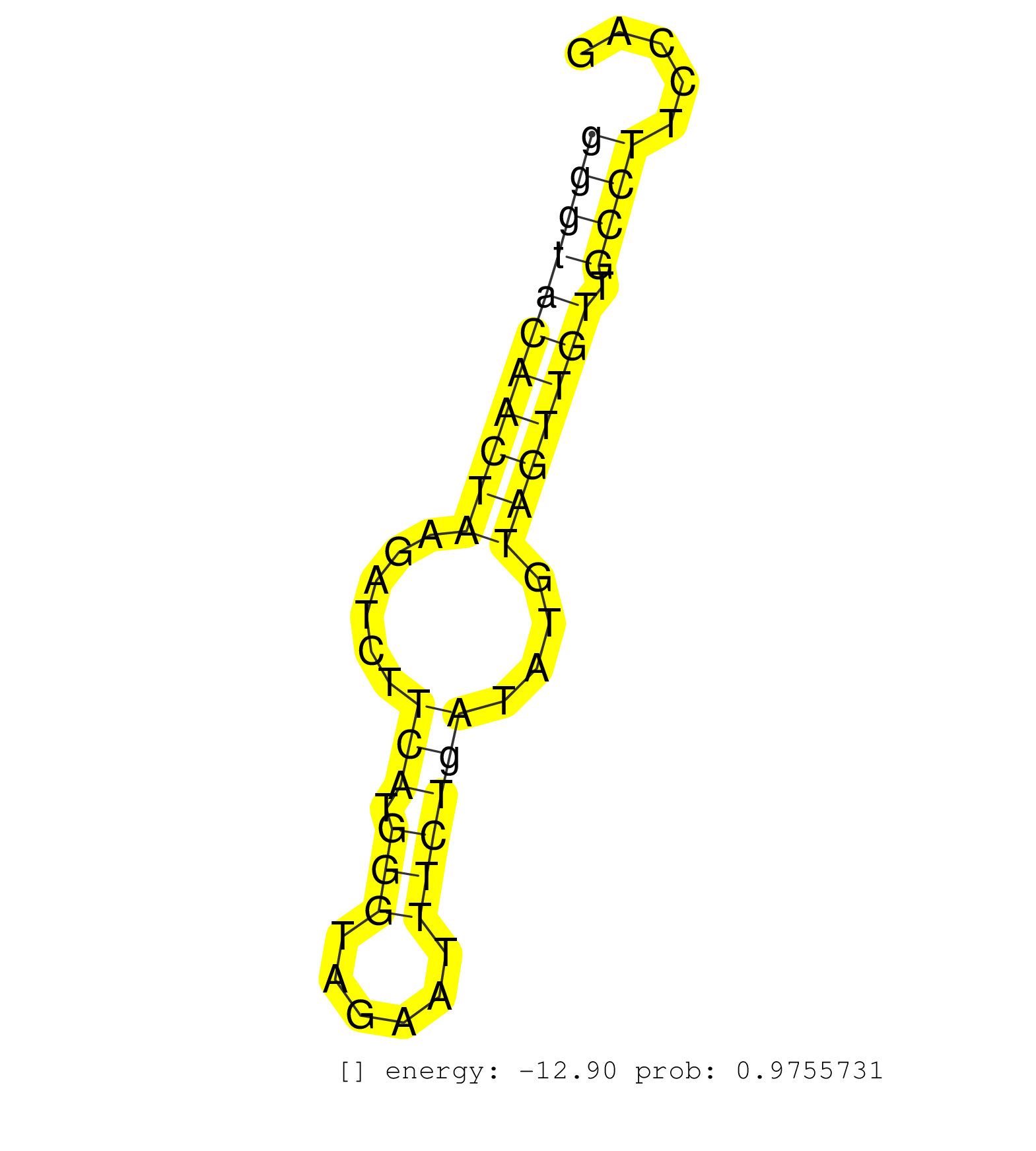
| ACTAGTGGTACCGAGTTTTGTTTCACTTTGTTTGTTTACCCATGGATGTGGTGTAATCGAGAAGACCATGTCTCAGGGCCCATCCAGCAGCCATTTAGTCCATCCTTTGTCATCATAAATCTGGTTAACTACTGGGTAGGTGGGGGTACAACTAAGATCTTCATGGGTAGAATTTCTGATATGTAGTTGTTGCCTTCCAGAGAAGTCATGGAGAGAGATGCAAAGGACTACGCCGACTCTATTCATGCAA ............................................................................................................................................((((((((((((((......(((.(((......))))))....))))))).)))).)))................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189782 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR040010(GSM532895) G529N. (cervix) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR191467(GSM715577) 115genomic small RNA (size selected RNA from . (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR040033(GSM532918) G603T. (cervix) | SRR040012(GSM532897) G648N. (cervix) | SRR189786 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR040015(GSM532900) G623T. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM532880(GSM532880) G659T. (cervix) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................CAACTAAGATCTTCATGGGTAGAATTTCT......................................................................... | 29 | 1 | 32.00 | 32.00 | 17.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CATGGAGAGAGATGCAAAGG........................ | 20 | 1 | 5.00 | 5.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATATGTAGTTGTTGCCTTCCAGT................................................. | 23 | 1 | 5.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGAGAGATGCAAAGGACTA.................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATATGTAGTTGTTGCCTTCCAG.................................................. | 22 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAGAAGTCATGGAGAGAG................................. | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................ATGGAGAGAGATGCAAAGGA....................... | 20 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CATGGAGAGAGATGCAAAGGT....................... | 21 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................GAAGTCATGGAGAGAGATG.............................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAGAAGTCATGGAGAGAGATGCAAAG......................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AAGTCATGGAGAGAGATGCAAAGGA....................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................ATGGAGAGAGATGCAAAGG........................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GTCATGGAGAGAGATGCAAAGT........................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATATGTAGTTGTTGCCTTCCAGA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AGAAGTCATGGAGAGAGATG.............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AGGACTACGCCGACTCTATTCATG... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GCAAAGGACTACGCCGAC............. | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............AGTTTTGTTTCACTTTACA.......................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................ACTACGCCGACTCTATTCACGCA. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................TGCAAAGGACTACGCC................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CAAAGGACTACGCCGAC............. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GTCATGGAGAGAGATGCAAAG......................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAGAAGTCATGGAGAGA.................................. | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GGTGGGGGTACAACTCTG.............................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................GAGAGAGATGCAAAGGACTACGCCGA.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................TAGTCCATCCTTTGTCATCAAAAT................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................CATGGAGAGAGATGCAAAGGACT..................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TATGTAGTTGTTGCCTTCCAGCT................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................AGATGCAAAGGACTACGG................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................GAGAGATGCAAAGGACTA.................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CATGGAGAGAGATGCAAAGGTT...................... | 22 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCATGGAGAGAGATGCAAAGGACTACGCCCACT............ | 33 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................AAGGACTACGCCGACTCTATTCACGC.. | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................GAGATGCAAAGGACTACGCCGACTCTATTCATG... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CATGGAGAGAGATGCAAA.......................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................GTCATGGAGAGAGATGCAAAGG........................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AGAAGTCATGGAGAGAGAT............................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TATGTAGTTGTTGCCTTCCAGT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................GAGATGCAAAGGACTACGCCGACT............ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GAGAGATGCAAAGGACTAC................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GCAAAGGACTACGCCGACT............ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................ACAACTAAGATCTTCATGGGTAGAATTT........................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGAGATGCAAAGGACTACGC................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GAAGTCATGGAGAGAGATGC............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TTTTGTTTCACTTTGTTTTAGG..................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................CATGGAGAGAGATGCAAAGGA....................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGAGATGCAAAGGACTACG.................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGTAGAATTTCTGATATGTA................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TTTCTGATATGTAGTTGTTGCCTT...................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TTCTGATATGTAGTTTGC........................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................AGAAGTCATGGAGAGAGATGCAA........................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CAACTAAGATCTTCATGGGTAGAATTTAT......................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................GCAAAGGACTACGCCGACTCTATTCATG... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGGGTAGAATTTCTGATATG................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAGAAGTCATGGAGAG................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ACTAGTGGTACCGAGTTTTGTTTCACTTTGTTTGTTTACCCATGGATGTGGTGTAATCGAGAAGACCATGTCTCAGGGCCCATCCAGCAGCCATTTAGTCCATCCTTTGTCATCATAAATCTGGTTAACTACTGGGTAGGTGGGGGTACAACTAAGATCTTCATGGGTAGAATTTCTGATATGTAGTTGTTGCCTTCCAGAGAAGTCATGGAGAGAGATGCAAAGGACTACGCCGACTCTATTCATGCAA ............................................................................................................................................((((((((((((((......(((.(((......))))))....))))))).)))).)))................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189782 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR040010(GSM532895) G529N. (cervix) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR191467(GSM715577) 115genomic small RNA (size selected RNA from . (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR040033(GSM532918) G603T. (cervix) | SRR040012(GSM532897) G648N. (cervix) | SRR189786 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR040015(GSM532900) G623T. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM532880(GSM532880) G659T. (cervix) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................CCATGTCTCAGGGCCCCG....................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................CCATGTCTCAGGGCCGCGA...................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................CTCTCTCCATGACTTCT................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ...................................................................................GACTAAATGGCTGCTGG...................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ........................................................................................................................................................................................................TCTCTCCATGACTTCT.................................. | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |