| (4) AGO2.ip | (2) B-CELL | (1) BRAIN | (8) BREAST | (18) CELL-LINE | (4) CERVIX | (1) KIDNEY | (8) LIVER | (1) OTHER | (9) SKIN | (1) XRN.ip |
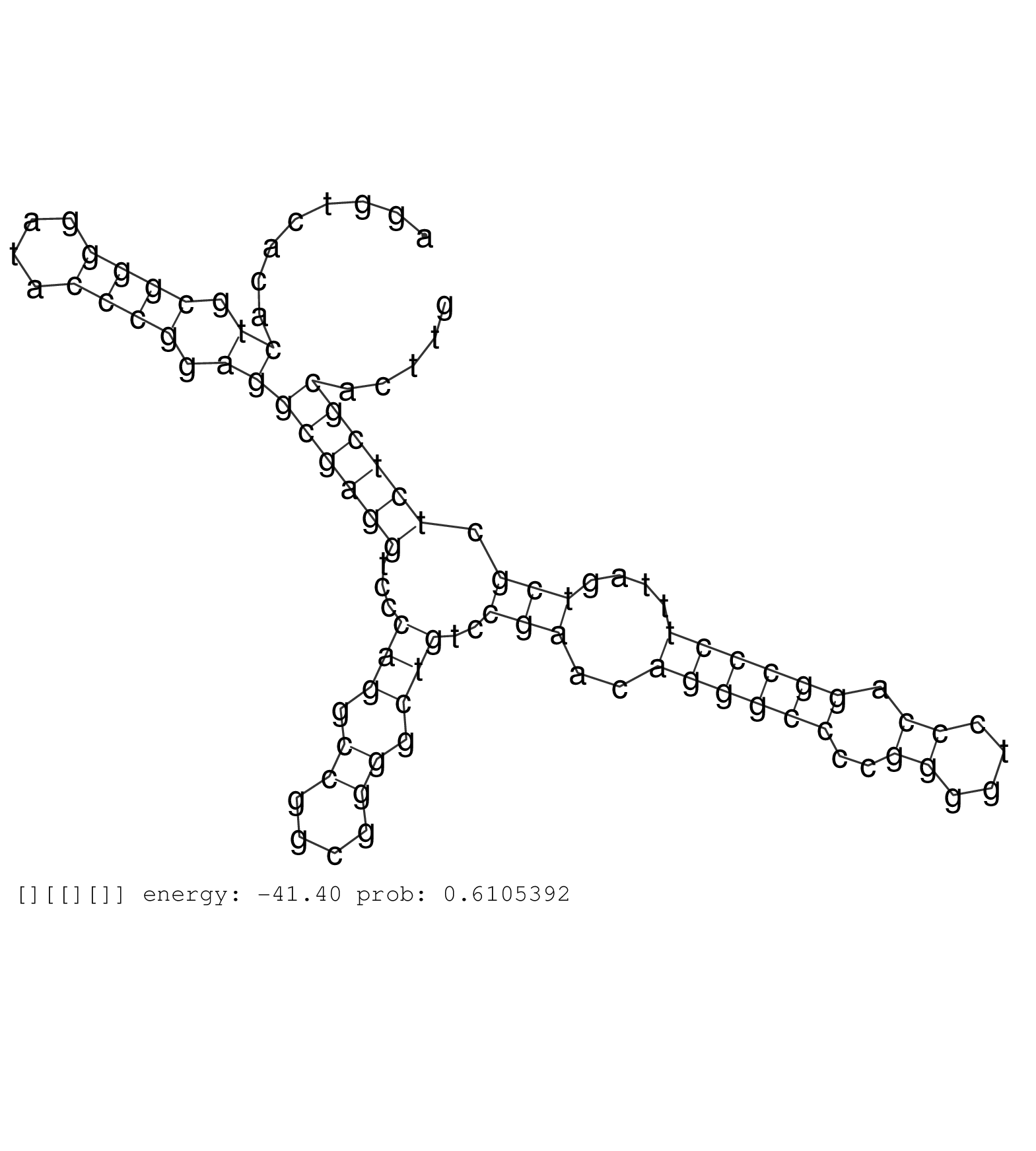
| AGCACAGTGCGGCCCCTGGACCACCTCCCAGTTCCCCAGGACCTCCTGAGGTAAGGTCGGGGCATGGGTCAGGATGACAGAGTGAGGCCGCTTCTCCCAGCCCCAGCATCATTTGCACATTGAGATGCAACAAAATAATCCCTGCCTCTTGAGACTGTGGTAAAAACTCACTGAGTCACACATATGCCAGCCCTAAGCTTAGAGTAAGGGTGGCTTTCTAGTGCTGGTAATAGAGAATGGAAACACCAGT ...................................................................(((.....))).((((...(((((.((((.........(((.....))).....((((.(((.............))).))))))))..)))))....))))................................................................................. ..............................................................63.........................................................................................................170.............................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | TAX577745(Rovira) total RNA. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM956925PazD5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM956925F181A(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444054(SRX128902) Sample 14cDNABarcode: AF-PP-342: ACG CTC TTC . (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR040018(GSM532903) G701N. (cervix) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR189787 | SRR040028(GSM532913) G026N. (cervix) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | TAX577590(Rovira) total RNA. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | GSM956925AGO2Paz8(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR343335 | TAX577579(Rovira) total RNA. (breast) | GSM956925Ago2(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | TAX577738(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................GTCAGGATGACAGAGCGGT.................................................................................................................................................................... | 19 | 16.00 | 0.00 | 1.00 | - | 3.00 | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| ...................................................................GTCAGGATGACAGAGTGGT.................................................................................................................................................................... | 19 | 13.00 | 0.00 | 4.00 | - | - | 1.00 | - | - | - | - | 2.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGCGG..................................................................................................................................................................... | 18 | 11.00 | 0.00 | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ...................................................................GTCAGGATGACAGAGTGGTC................................................................................................................................................................... | 20 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGTGG..................................................................................................................................................................... | 18 | 4.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................TTCCCCAGGACCTCCACCA........................................................................................................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ...................................................................GTCAGGATGACAGAGAGG..................................................................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ....................................................................TCAGGATGACAGAGTGAGGCCGC............................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GTCAGGATGACAGAGAGT..................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CTGCCTCTTGAGACTGTGGTAAAAACTCACTG............................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GTCAGGATGACAGAGTATTC................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGCAGG.................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................GTCGGGGCATGGGTCAGGATGACAGAGTGAGGCC................................................................................................................................................................. | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GTCAGGATGACAGAGAGGT.................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGTGTAC................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................ACCTCCTGAGGTAAGATGG............................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGTAG..................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGTGGTA................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................TGAGGCCGCTTCTCCGGG...................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGTGTT.................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ....................................................................TCAGGATGACAGAGTGGT.................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GTCAGGATGACAGAGCGGA.................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................AGCCCTAAGCTTAGAGTAAGG......................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TTGAGACTGTGGTAAAAA.................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCACAGTGCGGCCCCTGGACCACCTCCCAGTTCCCCAGGACCTCCTGAGGTAAGGTCGGGGCATGGGTCAGGATGACAGAGTGAGGCCGCTTCTCCCAGCCCCAGCATCATTTGCACATTGAGATGCAACAAAATAATCCCTGCCTCTTGAGACTGTGGTAAAAACTCACTGAGTCACACATATGCCAGCCCTAAGCTTAGAGTAAGGGTGGCTTTCTAGTGCTGGTAATAGAGAATGGAAACACCAGT ...................................................................(((.....))).((((...(((((.((((.........(((.....))).....((((.(((.............))).))))))))..)))))....))))................................................................................. ..............................................................63.........................................................................................................170.............................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | TAX577745(Rovira) total RNA. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM956925PazD5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM956925F181A(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444054(SRX128902) Sample 14cDNABarcode: AF-PP-342: ACG CTC TTC . (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR040018(GSM532903) G701N. (cervix) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR189787 | SRR040028(GSM532913) G026N. (cervix) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | TAX577590(Rovira) total RNA. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | GSM956925AGO2Paz8(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR343335 | TAX577579(Rovira) total RNA. (breast) | GSM956925Ago2(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | TAX577738(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................CTTCTCCCAGCCCCAGTCTG............................................................................................................................................ | 20 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .............................................................GCATGGGTCAGGATGAAA........................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................AGATGCAACAAAATAGAC.............................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................TTCTCCCAGCCCCAGTCTG............................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................CAACAAAATAATCCCTGT......................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ............................................CCGACCTTACCTCAGG.............................................................................................................................................................................................. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGCGGCCCCTGGACCGGT................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GGCTGGGAGAAGCGGCCTCACTCTGT.................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................CTGAGGTAAGGTCGGTTGG.......................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |