| (1) AGO2.ip | (7) B-CELL | (2) BREAST | (21) CELL-LINE | (1) CERVIX | (1) HEART | (2) HELA | (4) LIVER | (1) OTHER | (1) RRP40.ip | (12) SKIN | (1) TESTES | (1) XRN.ip |
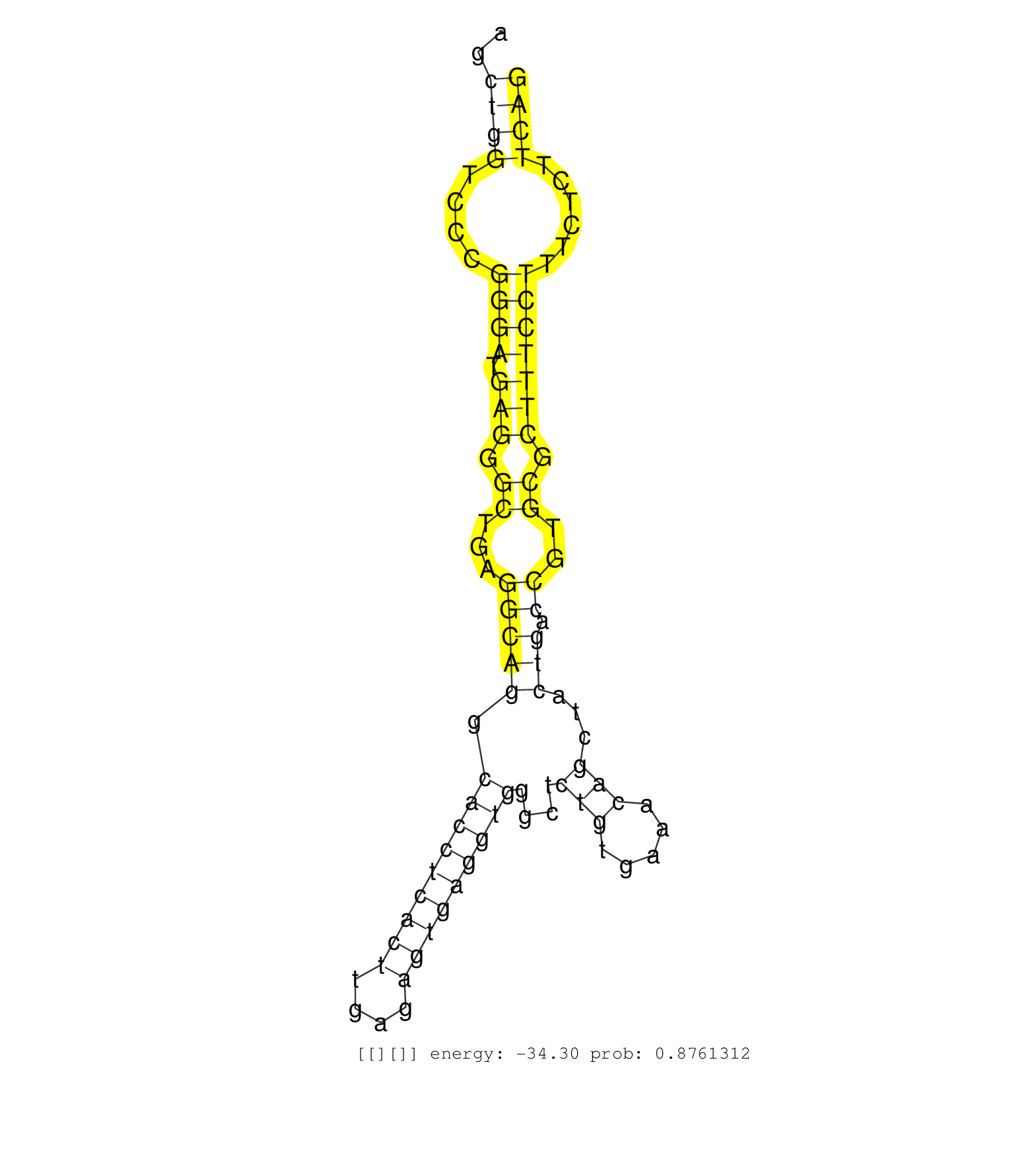
| GCCCAGCCTAGCACCATTGCCACCAGCATGCGGGATTCCCTCATTGACAGGTACCTGGGGCCTCTCCTTTCTTCTGCTCAGGGTTGGGGGTTAACTTGCATTTTGAGAGTATGGGGAAAAGATGTCATCTAGAATGGGAGCCTACCAGTTTGTTCTGGTAGCTTCTCGAGTGTGAGAGCATATCTGTACACTGCTGAGCAGATAAGAGCCAGCATATTCAAAAAATATTTCTTGAGCATCTGCTAATCCTCATCTTTGTGGTTTTTCAGGGCTAGGATACTAGAGCAGTAGCTGCTTGAGCAGCTTGTCAGAGCTGGTCCCGGGATGAGGGCTGAGGCAGGCACCTCACTTGAGAGTGAGGTGGGCTCTGTGAAACAGCTACTGACCGTGCGCTTTCCTTTCTCTTCAGCCTCACCTGAGTCACCTTCCAAGTTGTTCCATGGGCTCCTGGCTCTGGAC .........................................................................................................................................................................................................................................................................................................................((((....((((.(((.((...(((((.(((((((((....)))))))))....(((.....)))...))).))..)).)))))))......)))).................................................. .......................................................................................................................................................................................................................................................................................................................312..............................................................................................409................................................ | Size | Perfect hit | Total Norm | Perfect Norm | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037935(GSM510473) 293cand3. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189782 | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR553576(SRX182782) source: Testis. (testes) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR189785 | SRR038861(GSM458544) MM466. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR189783 | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR343336 | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR029127(GSM416756) A549. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................................................................................................................GTCCCGGGATGAGGGCTGAGG.......................................................................................................................... | 21 | 1 | 13.00 | 13.00 | 2.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................GTCCCGGGATGAGGGCTGAGGC......................................................................................................................... | 22 | 1 | 12.00 | 12.00 | 6.00 | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................GTCCCGGGATGAGGGCTGAGGCA........................................................................................................................ | 23 | 1 | 10.00 | 10.00 | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................GTCCCGGGATGAGGGCTGAGGCAA....................................................................................................................... | 24 | 1 | 7.00 | 10.00 | - | - | - | - | - | 1.00 | - | 3.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................................CGTGCGCTTTCCTTTCTCTTCAGT................................................. | 24 | 1 | 6.00 | 2.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................GTCCCGGGATGAGGGCTGAGGCAG....................................................................................................................... | 24 | 1 | 6.00 | 6.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................................................................................................................................GTCCCGGGATGAGGGCTGAG........................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................CGGGATGAGGGCTGAGGCAGGCAC................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................TGAGAGTGAGGTGGGCTCTGTGT...................................................................................... | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................CGGGATTCCCTCATTGACAGCCTC..................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................................................................................................................................................................................CGTGCGCTTTCCTTTCTCTTCAG.................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CTAGCACCATTGCCATCAG................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................CTAGAATGGGAGCCTACCAGTTT.................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................GTCACCTTCCAAGTTGTTC..................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TGTACACTGCTGAGCAGATAAGA............................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................ATGCGGGATTCCCTCATTGACAGCCT...................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................CTTCTGCTCAGGGTTA..................................................................................................................................................................................................................................................................................................................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................................................GGTTTTTCAGGGCTAGGATACTAGAGC............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................GTTGTTCCATGGGCTCCTGG........ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................AGGGCTAGGATACTAGAG.............................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................................................TGCGCTTTCCTTTCTCTTCAGT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............CCATTGCCACCAGCATGCGG.......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CTAGCACCATTGCCATCCG................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......CCTAGCACCATTGCCACCAGC................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................GTTGTTCCATGGGCTC............ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................CACCTTCCAAGTTGTTCCATGGGCTCCTGGC....... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................TCCCGGGATGAGGGCTGAGG.......................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................ACCAGCATGCGGGATTCCC................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................TGAGAGCATATCTGTTGG............................................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .....................ACCAGCATGCGGGAT....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................AAGTTGTTCCATGGGCTC............ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGGCTCCTGGCTCTGGAC | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................CAGCCTCACCTGAGTCACCTTCCAAG........................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................CCAAGTTGTTCCATGGGCTC............ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................ATGAGGGCTGAGGCAG....................................................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| GCCCAGCCTAGCACCATTGCCACCAGCATGCGGGATTCCCTCATTGACAGGTACCTGGGGCCTCTCCTTTCTTCTGCTCAGGGTTGGGGGTTAACTTGCATTTTGAGAGTATGGGGAAAAGATGTCATCTAGAATGGGAGCCTACCAGTTTGTTCTGGTAGCTTCTCGAGTGTGAGAGCATATCTGTACACTGCTGAGCAGATAAGAGCCAGCATATTCAAAAAATATTTCTTGAGCATCTGCTAATCCTCATCTTTGTGGTTTTTCAGGGCTAGGATACTAGAGCAGTAGCTGCTTGAGCAGCTTGTCAGAGCTGGTCCCGGGATGAGGGCTGAGGCAGGCACCTCACTTGAGAGTGAGGTGGGCTCTGTGAAACAGCTACTGACCGTGCGCTTTCCTTTCTCTTCAGCCTCACCTGAGTCACCTTCCAAGTTGTTCCATGGGCTCCTGGCTCTGGAC .........................................................................................................................................................................................................................................................................................................................((((....((((.(((.((...(((((.(((((((((....)))))))))....(((.....)))...))).))..)).)))))))......)))).................................................. .......................................................................................................................................................................................................................................................................................................................312..............................................................................................409................................................ | Size | Perfect hit | Total Norm | Perfect Norm | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037935(GSM510473) 293cand3. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189782 | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR553576(SRX182782) source: Testis. (testes) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR189785 | SRR038861(GSM458544) MM466. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR189783 | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR343336 | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR029127(GSM416756) A549. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................................................................................................................................................GAGGTGGGCTCTGTGAACCG.................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................TCAAGAAATATTTTTTGAATATGCTGGC................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................TGGAACAACTTGGAAGGTGACTCAGG................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................GCATCTGCTAATCCTCATA............................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................AATATTTCTTGAGCAGATG......................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................ATCTGTACACTGCTGAGCCGC................................................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................TGGAAGGTGACTCAGGTGAGGCTGAA............................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................TTGGAAGGTGACTCAGGTGAGGCT............................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................TGACTCAGGTGAGGC.................................... | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |