| (1) AGO1.ip | (2) AGO2.ip | (1) B-CELL | (3) BRAIN | (5) BREAST | (27) CELL-LINE | (5) CERVIX | (7) HEART | (1) HELA | (1) KIDNEY | (4) LIVER | (2) OTHER | (24) SKIN | (1) TESTES | (1) XRN.ip |
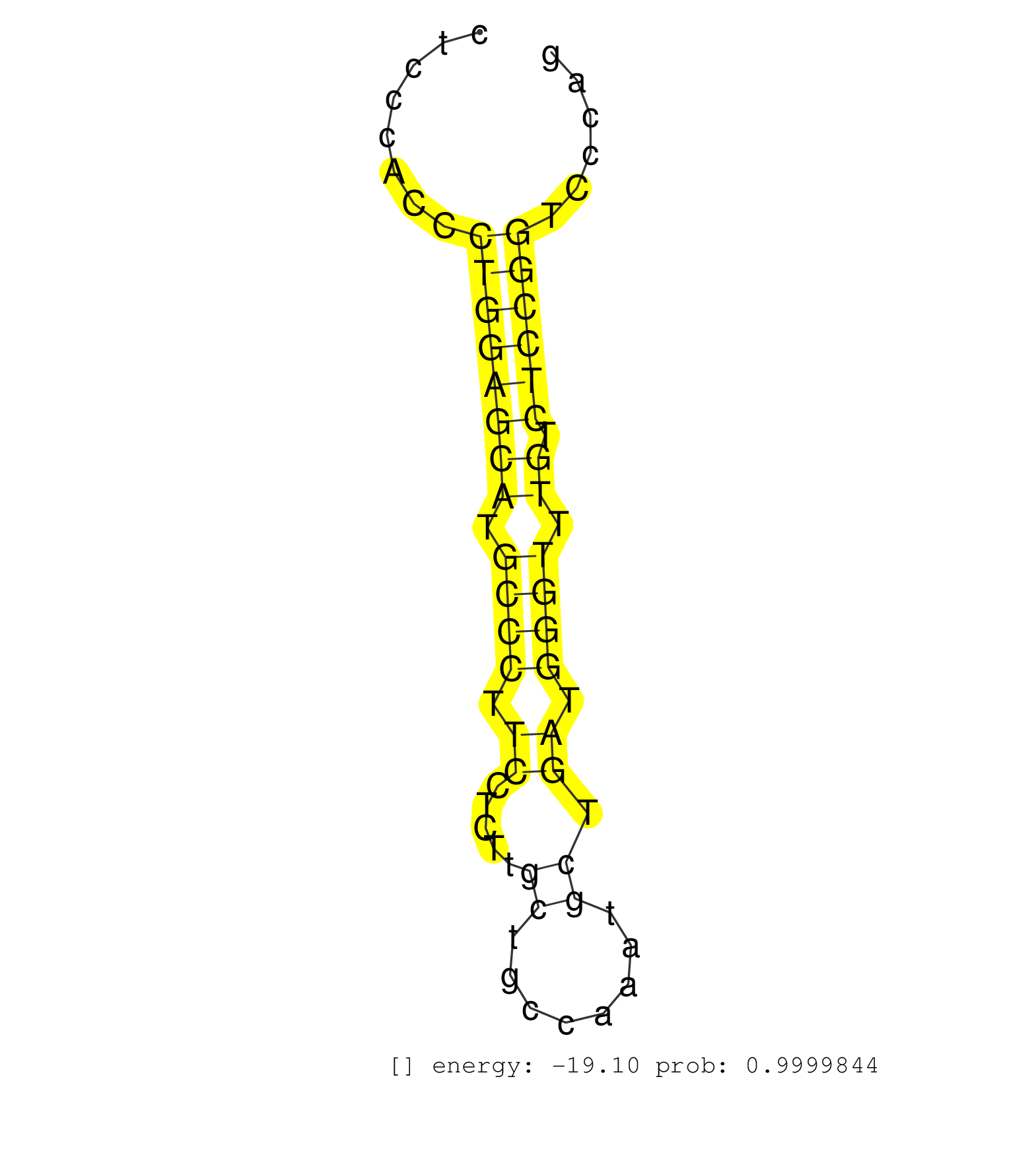
| TGTCCTCTGAGCATCTGTAGCAGTAGTGTGGCAGGAGGGGAAGATGGGCAAGGCCGAGAGAGCCCTTTCCTGTTCAGGGTTCTTGGCAGGGATGGGACCGGTGGGGCCTGGGCCCTGGCTGACTCCGGCCTGTGACTCCCACCCTGGAGCATGCCCTTCCTCTTGCTGCCAAATGCTGATGGGTTTGTCTCCGGTCCCAGAGCACCAAGCTATGGATCATCATGGAGTACCTGGGCGGCGGCTCAGCACT ...............................................................................................................................................((((((((.((((.((.....((........)).)).)))).)).))))))........................................................ .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR040039(GSM532924) G531T. (cervix) | SRR029127(GSM416756) A549. (cell line) | SRR189786 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR040011(GSM532896) G529T. (cervix) | SRR189783 | SRR037937(GSM510475) 293cand2. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343334 | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR444065(SRX128913) Sample 22cDNABarcode: AF-PP-335: ACG CTC TTC . (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR029129(GSM416758) SW480. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR191449(GSM715559) 144genomic small RNA (size selected RNA from . (breast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR038858(GSM458541) MEL202. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR191456(GSM715566) 182genomic small RNA (size selected RNA from . (breast) | SRR029126(GSM416755) 143B. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR040032(GSM532917) G603N. (cervix) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | SRR040043(GSM532928) G428T. (cervix) | SRR038853(GSM458536) MELB. (cell line) | SRR444066(SRX128914) Sample 23cDNABarcode: AF-PP-339: ACG CTC TTC . (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................ACCCTGGAGCATGCCCTTCCTC........................................................................................ | 22 | 1 | 10.00 | 10.00 | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGGTC...................................................... | 20 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGGGTTTGTCTCCGGTCCCAGA................................................. | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GATCATCATGGAGTAACT.................. | 18 | 5.00 | 0.00 | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................ACCCTGGAGCATGCCCTTCCT......................................................................................... | 21 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GACTCCCACCCTGGAGCC................................................................................................... | 18 | 3.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................ACCCTGGAGCATGCCCTTCCTCT....................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGGTCT..................................................... | 21 | 1 | 2.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CAGGGATGGGACCGGGAG.................................................................................................................................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGGTCCCATC................................................. | 25 | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TGGGTTTGTCTCCGGTCCCAGT................................................. | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGGT....................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATGGATCATCATGGAGTACCTGGGCGG............ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACCCTGGAGCATGCCCTT............................................................................................ | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGGTCCC.................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................GAGAGAGCCCTTTCCTGTTA............................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................CATGGAGTACCTGGGCGGCGGCTCAGC... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CATGGAGTACCTGGGCG............. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGAGCATCTGTAGCAGTAGTGTGGC.......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGTCCC..................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................ATCATCATGGAGTACCTGGGCGG............ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GACTCCCACCCTGGAGCCTCC................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GAGCACCAAGCTATGGATCATCATGGA........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................CATCATGGAGTACCTGGGCGGCGGCT....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................TGGGCGGCGGCTCAGCACT | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GAGTACCTGGGCGGCGGCTC...... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATGGGTTTGTCTCCGGTCCCAGA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................GTGGCAGGAGGGGAACGGC............................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................ATCATCATGGAGTACCTGGGCGGCGGCT....... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CCCTGGAGCATGCCCCC............................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................TGGGCCCTGGCTGACTGGGA.......................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................TCATCATGGAGTACCTGGGCGGCGGCT....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CAGGGATGGGACCGGGAA.................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................ATGGAGTACCTGGGCGGCG.......... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGAGTACCTGGGCGGCGG......... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GACTCCCACCCTGGAGCCTC................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............TGTAGCAGTAGTGTGCAT......................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCG......................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GACTCCCACCCTGGAGCCT.................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................TACCTGGGCGGCGGCATGG.... | 19 | 4 | 1.00 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CCCTGGAGCATGCCCTTCCTC........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CACCAAGCTATGGATCATCATGA......................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................GACTCCCACCCTGGAGCA................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CTGTTCAGGGTTCTTGGCAGGGATGGGACCGGT.................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACCCTGGAGCATGCCCTTGCTC........................................................................................ | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................TGGATCATCATGGAGTACCTGGGCGGCG.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................ATCATCATGGAGTACCTGGA............... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTCCCACCCTGGAGCACC................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................ACCTGGGCGGCGGCTTG..... | 17 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGGTCC..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGATGGGTTTGTCTCCGGACTC.................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................TATGGATCATCATGGAGTACCTGGGC.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CTGTAGCAGTAGTGTAGGG......................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................AGTACCTGGGCGGCGGCTCAGCACCGG | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTCCCACCCTGGAGC.................................................................................................... | 16 | 7 | 0.57 | 0.57 | - | - | - | - | - | - | - | - | - | - | - | 0.29 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - |
| ................................AGGAGGGGAAGATGGGC......................................................................................................................................................................................................... | 17 | 4 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTCCCACCCTGGAGCAT.................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTCCCACCCTGGAGCA................................................................................................... | 16 | 6 | 0.50 | 0.50 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - |
| ...................................................................................................................................................................................................................................TACCTGGGCGGCGGC........ | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....CTCTGAGCATCTGTAG...................................................................................................................................................................................................................................... | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - |
| ......................................................................................................................................ACTCCCACCCTGGAGCCC.................................................................................................. | 18 | 7 | 0.14 | 0.57 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTCCCACCCTGGAGCGT.................................................................................................. | 18 | 7 | 0.14 | 0.57 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| TGTCCTCTGAGCATCTGTAGCAGTAGTGTGGCAGGAGGGGAAGATGGGCAAGGCCGAGAGAGCCCTTTCCTGTTCAGGGTTCTTGGCAGGGATGGGACCGGTGGGGCCTGGGCCCTGGCTGACTCCGGCCTGTGACTCCCACCCTGGAGCATGCCCTTCCTCTTGCTGCCAAATGCTGATGGGTTTGTCTCCGGTCCCAGAGCACCAAGCTATGGATCATCATGGAGTACCTGGGCGGCGGCTCAGCACT ...............................................................................................................................................((((((((.((((.((.....((........)).)).)))).)).))))))........................................................ .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR040039(GSM532924) G531T. (cervix) | SRR029127(GSM416756) A549. (cell line) | SRR189786 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR040011(GSM532896) G529T. (cervix) | SRR189783 | SRR037937(GSM510475) 293cand2. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343334 | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR444065(SRX128913) Sample 22cDNABarcode: AF-PP-335: ACG CTC TTC . (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR029129(GSM416758) SW480. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR191449(GSM715559) 144genomic small RNA (size selected RNA from . (breast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR038858(GSM458541) MEL202. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR191456(GSM715566) 182genomic small RNA (size selected RNA from . (breast) | SRR029126(GSM416755) 143B. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR040032(GSM532917) G603N. (cervix) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | SRR040043(GSM532928) G428T. (cervix) | SRR038853(GSM458536) MELB. (cell line) | SRR444066(SRX128914) Sample 23cDNABarcode: AF-PP-339: ACG CTC TTC . (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................GTTCAGGGTTCTTGGTCTG................................................................................................................................................................ | 19 | 37.00 | 0.00 | 15.00 | - | 3.00 | - | - | - | - | - | - | - | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................GTTCAGGGTTCTTGGCTCTG............................................................................................................................................................... | 20 | 5.00 | 0.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................CTCCCACCCTGGAGCAAA................................................................................................. | 18 | 4.00 | 0.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTCCCACCCTGGAGCAAAA................................................................................................ | 20 | 7 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GCTCCAGGGTGGGAG.................................................................................................... | 15 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................CCTGTTCAGGGTTCTTGGAG.................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTCCCACCCTGGAGCAGAA................................................................................................ | 20 | 7 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GTTCAGGGTTCTTGGCATCTG.............................................................................................................................................................. | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................CTCCCACCCTGGAGCATGCCCTTCCTCTCC..................................................................................... | 30 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................AGAGAGCCCTTTCCTGTTTC.............................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................ACCCTGAACAGGAAAGGGCT.......................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTCCCACCCTGGAGAGA.................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................GTGGCAGGAGGGGAAGTA............................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................CTCGGCCTTGCCCATC................................................................................................................................................................................................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTCCCACCCTGGAGCACA................................................................................................. | 19 | 7 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGCTCTGGGACCGGA.............................................. | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |