| (1) AGO2.ip | (3) B-CELL | (2) BRAIN | (4) BREAST | (6) CELL-LINE | (1) CERVIX | (1) HEART | (5) LIVER | (6) OTHER | (4) SKIN | (1) UTERUS |
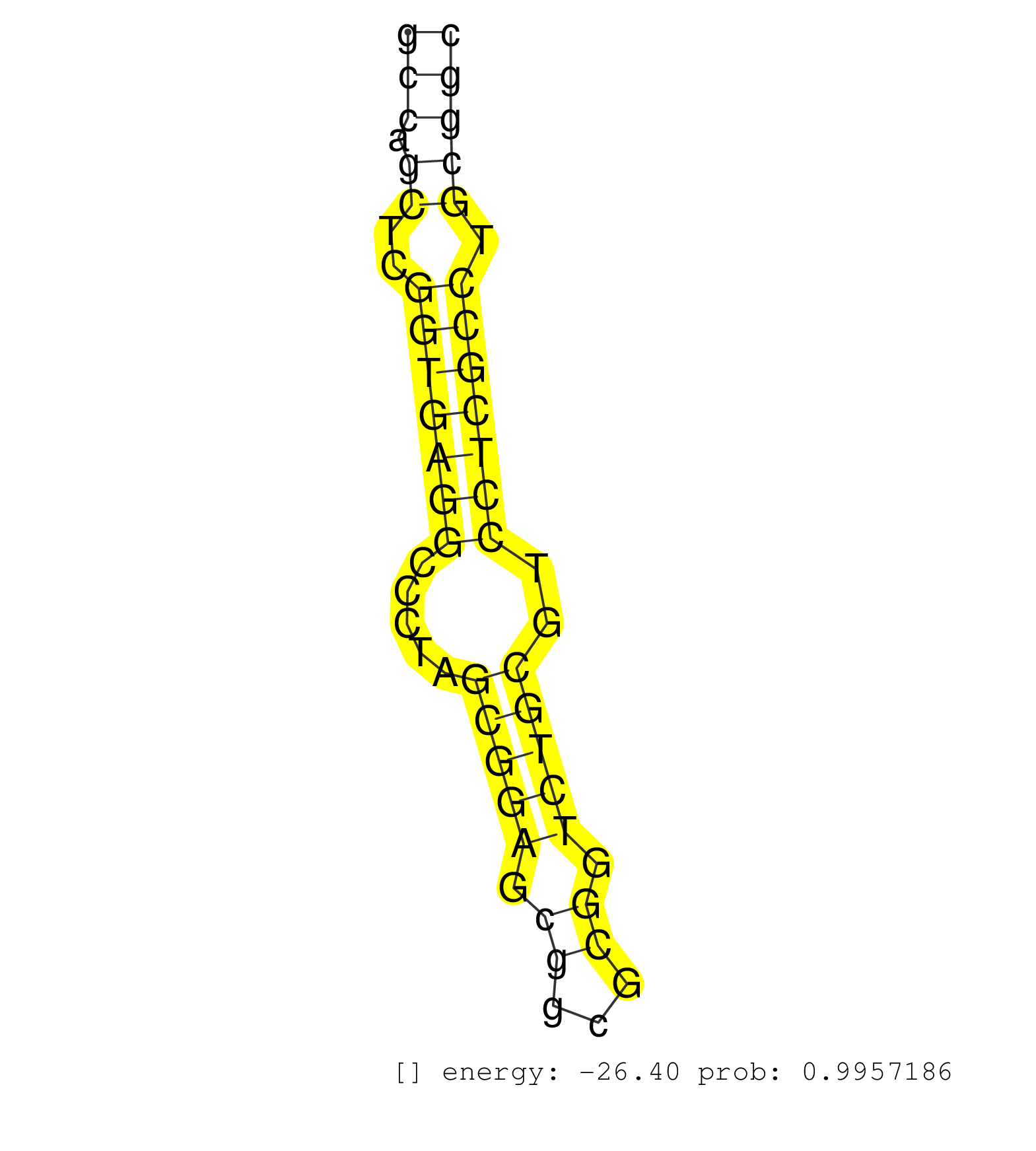
| AAAGATTGAAAAAGAATTGGAAAGGGGTAATTTGTTGGGCCATTCAGTGCTTTGCTGTCATTCATGGTGTCATCGTGTTGTGCAGCCACCACATAGCACTGAATAGTGAGGAGATGTTGTTTGCTGTGGACGTAGAAAACCATCTTGTCCCCACTTTGGAAAGTGCTGCTCGTGTCTCACAAAACTTACGCTTTTTGTAGGCATTCACTCATACGGCACAATATGATGAAGCAATTTCAGATTATTTCAG ...........................................................................................................((.((((((...(((((.......)))))....)))))).)).((.....))...(((..((....))..)))...................................................................... ..........................................................................................................107.......................................................................181................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189787 | SRR189786 | SRR189784 | SRR189782 | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343334 | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR040036(GSM532921) G243N. (cervix) | TAX577740(Rovira) total RNA. (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | TAX577743(Rovira) total RNA. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................AAAGTGCTGCTCGTGCAG......................................................................... | 18 | 19.00 | 0.00 | 8.00 | 2.00 | 4.00 | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................AAAGTGCTGCTCGTGCAGG........................................................................ | 19 | 17.00 | 0.00 | 7.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................GAAAGTGCTGCTCGTGCAG......................................................................... | 19 | 6.00 | 0.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................CATTCACTCATACGGCACAAT............................ | 21 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CATTCACTCATACGGCACAATATG......................... | 24 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................ACGGCACAATATGATGAA.................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................CAATTTCAGATTATTTCA. | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGGCATTCACTCATACGGCACAAAA........................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................CATTCACTCATACGGCACA.............................. | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CACTCATACGGCACAATATGATGA..................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AAAGTGCTGCTCGTGCCG......................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................AAAGGGGTAATTTGTAT..................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................CGGCACAATATGATG...................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................GAAAGTGCTGCTCGTGCAGG........................................................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................AAAGTGCTGCTCGTGAAGG........................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................ACGGCACAATATGATGAAGCAA................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................AGATGTTGTTTGCTGTGGACGTA.................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................................ATACGGCACAATATGATGAA.................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGTGGACGTAGAAAACTG............................................................................................................ | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGGCATTCACTCATACGGC................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAGTGAGGAGATGTTGTTT................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GAAAGTGCTGCTCGTGCCGG........................................................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................AGGAGATGTTGTTTGCTGTGG......................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CACAATATGATGAAGCAATTTCAG.......... | 24 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AAAGTGCTGCTCGTGCA.......................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................TACGGCACAATATGATGAAG................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................ACGGCACAATATGATGAAG................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................AAAGTGCTGCTCGTGC........................................................................... | 16 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................ACGGCACAATATGATGAAGC.................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................AGGAGATGTTGTTTGCTTT........................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................TTGCTGTGGACGTAGAAAACC............................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................ACGGCACAATATGATGAAGCA................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GAAAGTGCTGCTCGTTCAG......................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................AGCCACCACATAGCAGAC..................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ..................................................................................................................................................................................................................ATACGGCACAATATGATGAAGCA................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................AAGCAATTTCAGATTATTTCGGGA | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ..............................................................................................................................................................GAAAGTGCTGCTCGTGCGC......................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................TGTAGGCATTCACTCATACGGCACA.............................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGACGTAGAAAACC............................................................................................................. | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AATAGTGAGGAGATG...................................................................................................................................... | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| AAAGATTGAAAAAGAATTGGAAAGGGGTAATTTGTTGGGCCATTCAGTGCTTTGCTGTCATTCATGGTGTCATCGTGTTGTGCAGCCACCACATAGCACTGAATAGTGAGGAGATGTTGTTTGCTGTGGACGTAGAAAACCATCTTGTCCCCACTTTGGAAAGTGCTGCTCGTGTCTCACAAAACTTACGCTTTTTGTAGGCATTCACTCATACGGCACAATATGATGAAGCAATTTCAGATTATTTCAG ...........................................................................................................((.((((((...(((((.......)))))....)))))).)).((.....))...(((..((....))..)))...................................................................... ..........................................................................................................107.......................................................................181................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189787 | SRR189786 | SRR189784 | SRR189782 | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343334 | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR040036(GSM532921) G243N. (cervix) | TAX577740(Rovira) total RNA. (breast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | TAX577743(Rovira) total RNA. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................ACAATATGATGAAGCAATAG............. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .......GAAAAAGAATTGGAATGCA................................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |