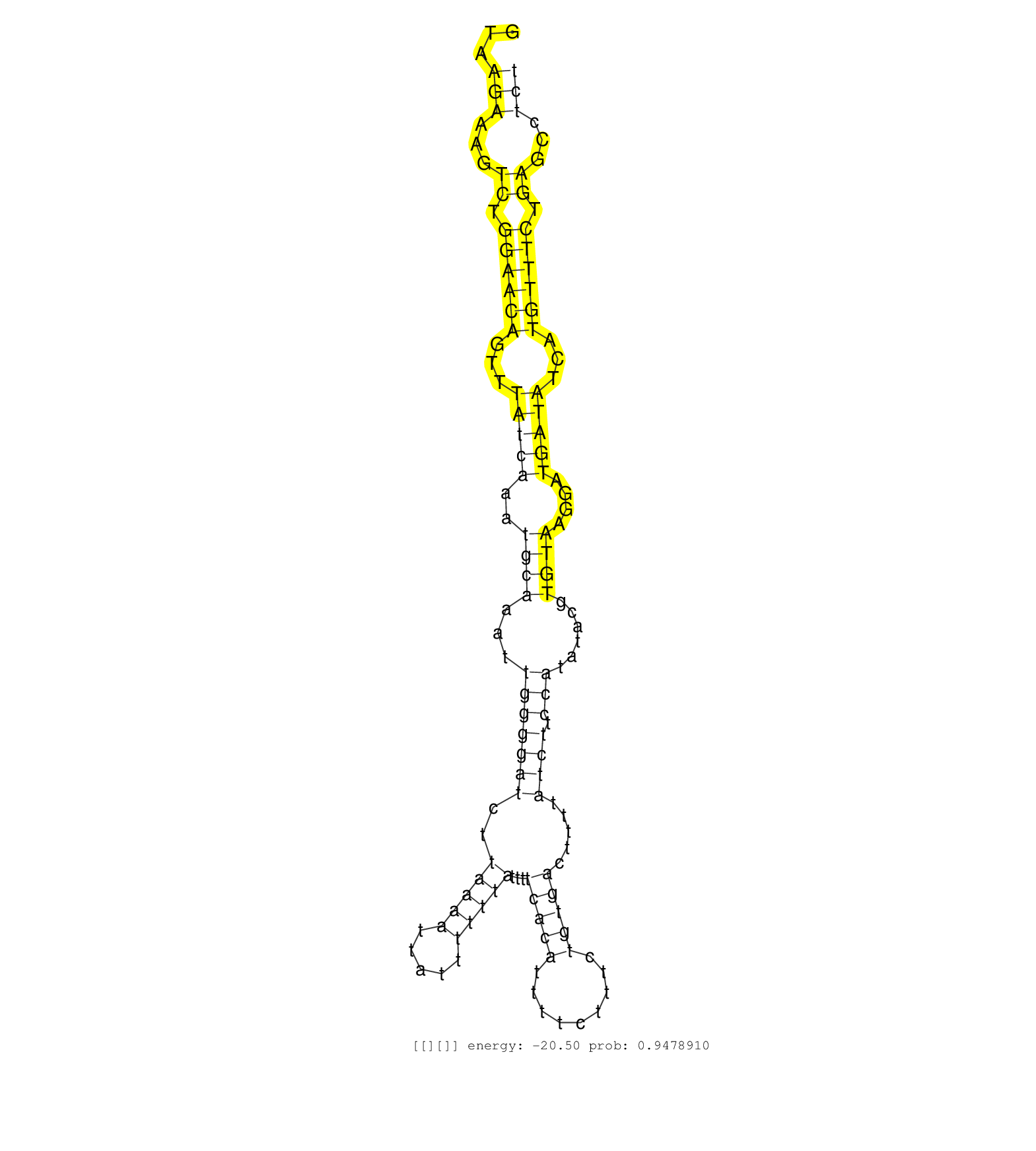
| (3) B-CELL | (1) BRAIN | (1) BREAST | (14) CELL-LINE | (1) HELA | (4) LIVER | (2) OTHER | (15) SKIN | (1) TESTES | (1) XRN.ip |
| CCACAGCTTTACCTCCCTGAAGGACAGACAAGGAACCCATAAAGATGCTGGTAAGAAAGTCTGGAACAGTTTATCAAATGCAAATTGGGGATCTTAAAATTATTTTTTATTTTCACATTTTCTTTCTGTGACTTTTATCTTCCATATACGTGTAAGGATGATATCATGTTTCTGAGCCTCTTATATATCCCTGGGGAGTCATCAGTCTGCTAAAGAGTGGCAAGAATCTTTGACTATTCACAATTCTGAA ....................................................((((((((.((((.((((((.........)))))).......(((((.....)))))..)))).)))))))).............................................................................................................................. ..................................................51...............................................................................132.................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189782 | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR038857(GSM458540) D20. (cell line) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................TGTAAGGATGATATCATGTTTCTGAGC......................................................................... | 27 | 1 | 44.00 | 44.00 | 8.00 | 4.00 | 4.00 | 3.00 | 3.00 | 3.00 | - | 2.00 | 2.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - |
| .......................................................................................................................................................GTAAGGATGATATCATGTTTCTGAGC......................................................................... | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TCCCTGAAGGACAGACAAGG......................................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................ATTTTCACATTTTCTTTTTA.......................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................AAGGAACCCATAAAGATGCTG........................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AATTATTTTTTATTTTCGGG..................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................GACAGACAAGGAACCCATAAAGATGCTG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CTCCCTGAAGGACAGACA............................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AAAGAGTGGCAAGAATCTTTGAC................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GAAGGACAGACAAGGAACCCATAAAGATA........................................................................................................................................................................................................... | 29 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GACAAGGAACCCATAAAGATGCTGAGAT.................................................................................................................................................................................................... | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................AAGGACAGACAAGGAACCCATAAAGATGC.......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................ATGCTGGTAAGAAAGTCTGGAACAGTT................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GAAAGTCTGGAACAGTGGTA................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................CAGACAAGGAACCCATAAAGATGCTG........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TGTAAGGATGATATCATGTTTCTGAG.......................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................TGTAAGGATGATATCATGTTTCT............................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................ATTTTTTATTTTCACGGGC.................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............CCTGAAGGACAGACAAGGAACCCATAAAGA............................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAAAGTCTGGAACAGTTTA................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................TGAAGGACAGACAAGGAA....................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...............................................................................................................TTCACATTTTCTTTCTG.......................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCACAGCTTTACCTCCCTGAAGGACAGACAAGGAACCCATAAAGATGCTGGTAAGAAAGTCTGGAACAGTTTATCAAATGCAAATTGGGGATCTTAAAATTATTTTTTATTTTCACATTTTCTTTCTGTGACTTTTATCTTCCATATACGTGTAAGGATGATATCATGTTTCTGAGCCTCTTATATATCCCTGGGGAGTCATCAGTCTGCTAAAGAGTGGCAAGAATCTTTGACTATTCACAATTCTGAA ....................................................((((((((.((((.((((((.........)))))).......(((((.....)))))..)))).)))))))).............................................................................................................................. ..................................................51...............................................................................132.................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189782 | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR038857(GSM458540) D20. (cell line) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................CATCAGTCTGCTAAATTGA................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................AAAGTCTGGAACAGTTGT................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |