| (2) B-CELL | (1) BRAIN | (2) BREAST | (4) CELL-LINE | (1) CERVIX | (6) HEART | (4) LIVER | (1) OTHER | (4) SKIN | (1) TESTES | (1) XRN.ip |
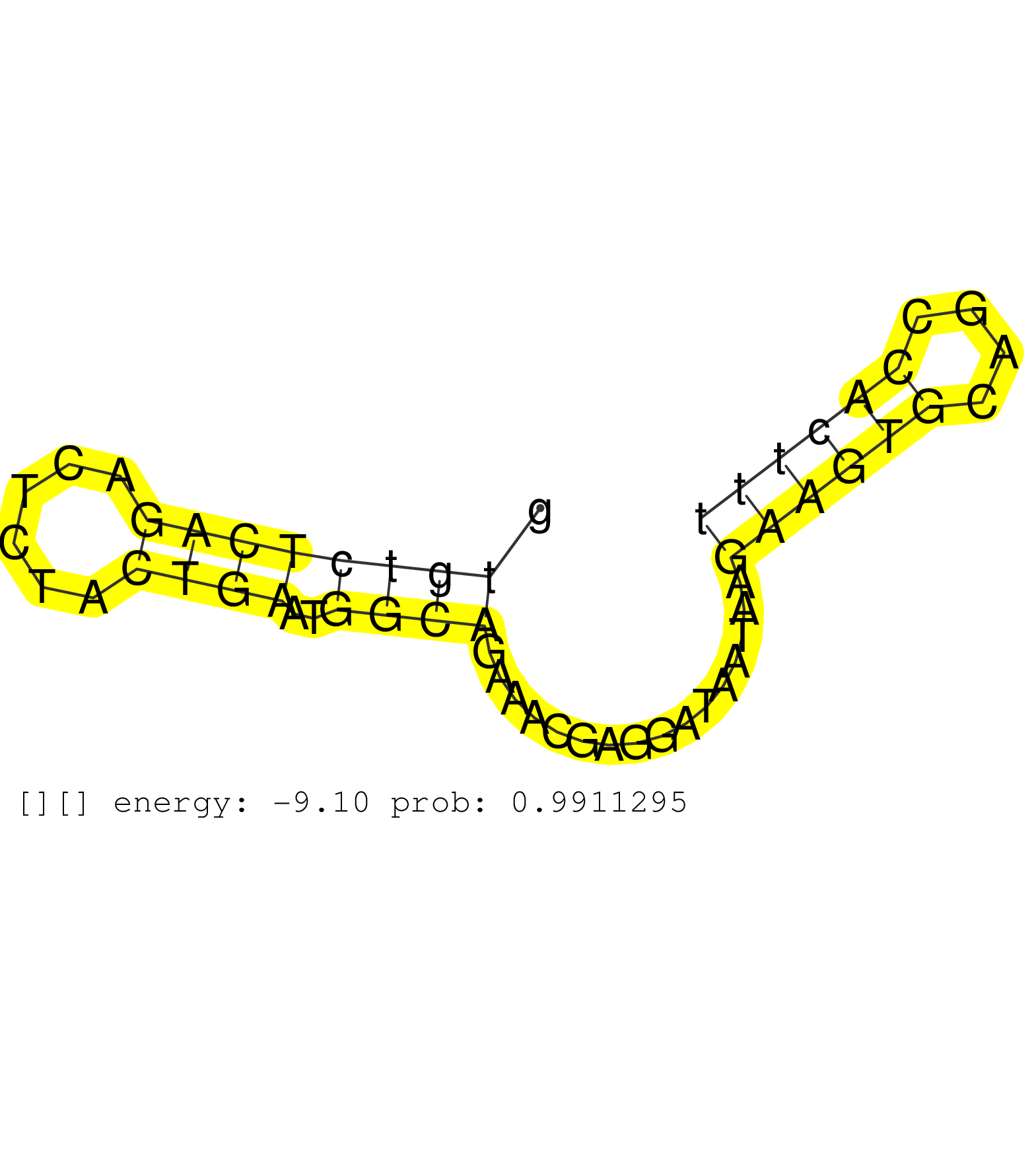
| GCTGTGGAGACCCACCTGCTCAGGAACCCTCACCTTGTTTCGGACTATAGGTAATTCATCAACTCTTCCTGAGGCTGGGTGGGTGGGAGGGAGTGAAGTGTCTCAGACTCTACTGAATGGCAGAAACGAGGATAATAAGAAGTGCAGCCACTTTACTGTCTCTGCCCACTTATCCAACTGCCAGATGATGTCACTTCATTCCTGTGAGCCCTTTAAATTCTTGAAACTTGTTTTCTTGCTTTCTTTTTCT .............................................................................(((((.((.(((.(((........)))...)))..(((.....))).....................)).))))).................................................................................................. .............................................................................78..........................................................................154.............................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | TAX577580(Rovira) total RNA. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR040037(GSM532922) G243T. (cervix) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189783 | SRR189782 | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR553576(SRX182782) source: Testis. (testes) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR189785 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................AGGATAATAAGAAGTGCAGCCA.................................................................................................... | 22 | 1 | 8.00 | 8.00 | 2.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AGGATAATAAGAAGTGCAGCC..................................................................................................... | 21 | 1 | 5.00 | 5.00 | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ACTGAATGGCAGAAACGAGGATAA................................................................................................................... | 24 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GTGGGAGGGAGTGAAAAA...................................................................................................................................................... | 18 | 3.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................CGAGGATAATAAGAAGTGCAGCC..................................................................................................... | 23 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AGGATAATAAGAAGTGCAGC...................................................................................................... | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CAGACTCTACTGAATGGCAGAAACG.......................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCAGACTCTACTGAATGGCAGAAACGA......................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CAGACTCTACTGAATGGCAGAAAAG.......................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................GAATGGCAGAAACGAGGATAATAAG............................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TTCATTCCTGTGAGCTCTG..................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................GAGGATAATAAGAAGTGCAGCCA.................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GAATGGCAGAAACGAGGATAATA................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CAGACTCTACTGAATGGCA................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCAGACTCTACTGAATGGC................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AATGGCAGAAACGAGGAT..................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AAACGAGGATAATAAGAAGTGCAGCCA.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TACTGAATGGCAGAAACGAGGATAATA................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................ATTCATCAACTCTTCG..................................................................................................................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................TGAATGGCAGAAACGAGGAT..................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GAATGGCAGAAACGAGGAT..................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GAATGGCAGAAACGAGGATAATAAGAAG............................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................CCCTCACCTTGTTTCACG.............................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................TGAGGCTGGGTGGGTGCCCG................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................CTACTGAATGGCAGAAACG.......................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CAGACTCTACTGAATGGCAGAAACGA......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................AGGAACCCTCACCTTGTTTCGGACTATAG........................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................AACGAGGATAATAAGAAGT........................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCAGACTCTACTGAATGGCAGAAACG.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................GTGGGTGGGAGGGAGTTTGA........................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................TGAATGGCAGAAACGAGGATA.................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GAAACGAGGATAATAAGAAGTGCAG....................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CTGAATGGCAGAAACGAGGATA.................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGAATGGCAGAAACGAGGATAA................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TTTTCTTGCTTTCTTTTCAAA | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ............................................................................................GTGAAGTGTCTCAGACTCTACTGAATGG.................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............CACCTGCTCAGGAACCCTCACCT....................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CTGAATGGCAGAAACGAGGATT.................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................AGGATAATAAGAAGTGCAG....................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TGTTTCGGACTATAG........................................................................................................................................................................................................ | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ..................................................................................................................................................................................................................................CTTGTTTTCTTGCTTTCTTT.... | 20 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .....................................................................................................................................................................................................................TAAATTCTTGAAACTT..................... | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| ....................................................................................................................................................................................................................................TGTTTTCTTGCTTTCTTT.... | 18 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - |
| GCTGTGGAGACCCACCTGCTCAGGAACCCTCACCTTGTTTCGGACTATAGGTAATTCATCAACTCTTCCTGAGGCTGGGTGGGTGGGAGGGAGTGAAGTGTCTCAGACTCTACTGAATGGCAGAAACGAGGATAATAAGAAGTGCAGCCACTTTACTGTCTCTGCCCACTTATCCAACTGCCAGATGATGTCACTTCATTCCTGTGAGCCCTTTAAATTCTTGAAACTTGTTTTCTTGCTTTCTTTTTCT .............................................................................(((((.((.(((.(((........)))...)))..(((.....))).....................)).))))).................................................................................................. .............................................................................78..........................................................................154.............................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | TAX577580(Rovira) total RNA. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR040037(GSM532922) G243T. (cervix) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189783 | SRR189782 | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR553576(SRX182782) source: Testis. (testes) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR189785 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......AGACCCACCTGCTCAACA................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................CTTATCCAACTGCCAGAA................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............CACCTGCTCAGGAACCCTCATGCC...................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................AAGTGCAGCCACTTTATT............................................................................................. | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................GTTTTCTTGCTTTCTGGGA.. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................GAGACAGTAAAGTGGCTGCA........................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................GAAGTGCAGCCACTTTAAG............................................................................................. | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................TTATCCAACTGCCAGAAA............................................................... | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................AAGTGCAGCCACTTTATTT............................................................................................ | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................ACCCTCACCTTGTTTCGGC.............................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |