| (1) AGO2.ip | (4) B-CELL | (11) BREAST | (15) CELL-LINE | (3) CERVIX | (1) HELA | (2) LIVER | (1) OTHER | (4) SKIN | (1) TESTES | (1) UTERUS |
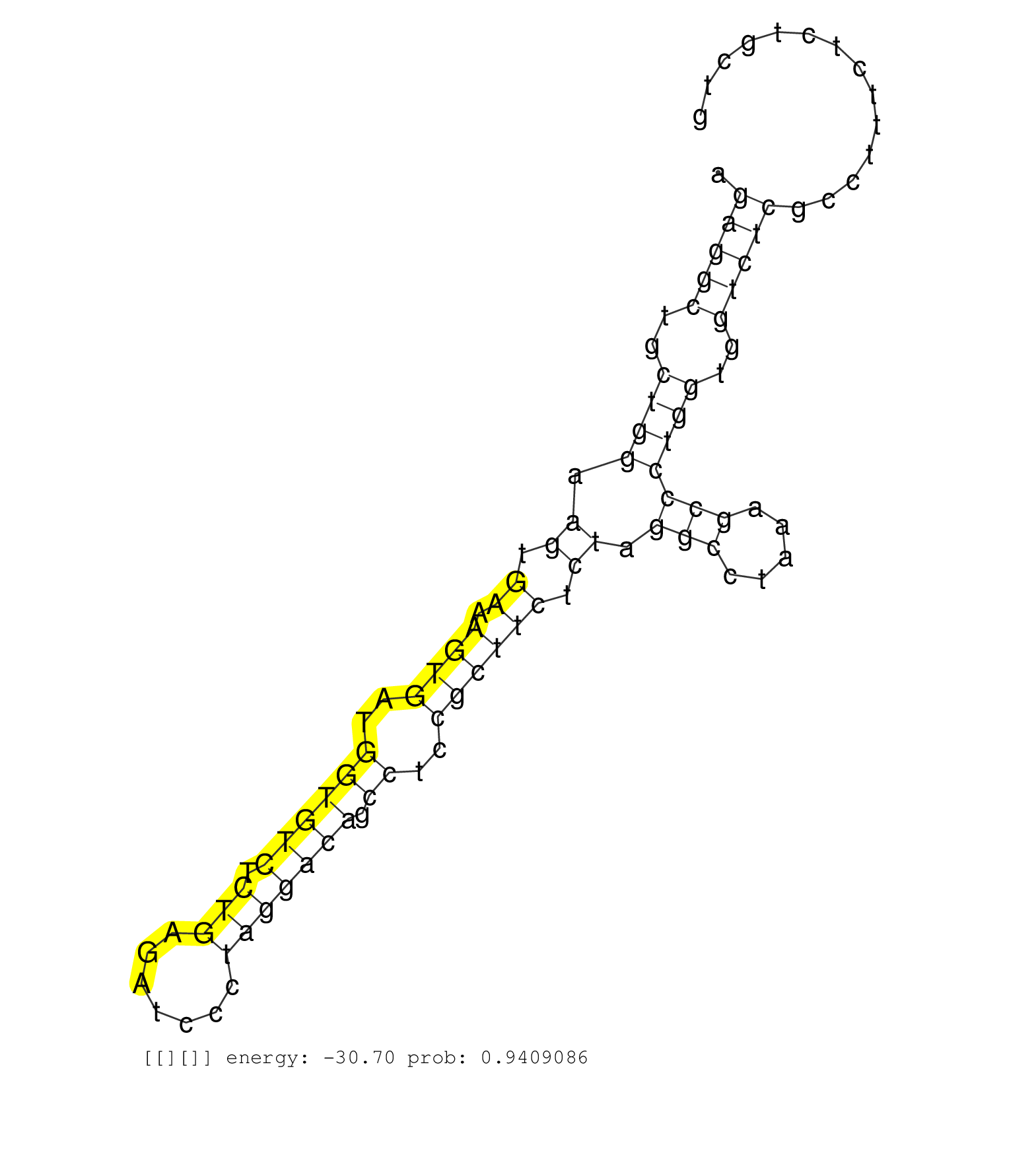
| TCATGTGCAAGAAAGACTTTCATCCTGCCTCCAAATCCAATATCAAAAAAGTGAGTAAACAGACCCGGGGAGGGATTAGGCCCACAGGGACCGCCTTCCCAGAGGCTGCTGGAAGTGAAAGTGATGGTGTCTCTGAGATCCCTAGGACAGCCTCCGCTTCTCTAGGCCTAAAGCCCTGGTGGTCTCGCCTTTCTCTGCTGAGCAGCTCGCTGACGCAGCGGGTCCTCAGCGTTTATTCTGCAACAGCGAG ...........................................................(((..((.(((((((....(((((...))).)).)))))))...))...)))..............((..((.....))..))............................................................................................................ ........................................................57...................................................................................142.......................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189782 | TAX577588(Rovira) total RNA. (breast) | SRR037938(GSM510476) 293Red. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191604(GSM715714) 74genomic small RNA (size selected RNA from t. (breast) | SRR040011(GSM532896) G529T. (cervix) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR040009(GSM532894) G727T. (cervix) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR040008(GSM532893) G727N. (cervix) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR553576(SRX182782) source: Testis. (testes) | SRR029132(GSM416761) MB-MDA231. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | TAX577746(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | TAX577739(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | TAX577738(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................GAAAGTGATGGTGTCTCTGAGA................................................................................................................ | 22 | 1 | 47.00 | 47.00 | 17.00 | 6.00 | 6.00 | 5.00 | - | - | 4.00 | 3.00 | - | - | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GTGCAAGAAAGACTTTCA.................................................................................................................................................................................................................................... | 18 | 1 | 4.00 | 4.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GACCCGGGGAGGGATTGACG......................................................................................................................................................................... | 20 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .........AGAAAGACTTTCATCCTGCCT............................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GCCCTGGTGGTCTCGCGGC........................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................AAGTGAAAGTGATGGTGGGAC..................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................AGTGATGGTGTCTCTGAGAT............................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGGAAGTGAAAGTGATGGTGT........................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAAAGTGATGGTGTCTCTGA.................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........AAGAAAGACTTTCATCCTG............................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CTGGAAGTGAAAGTGAGTTT.......................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................ACCCGGGGAGGGATTGAC.......................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................AGGCTGCTGGAAGTGAAAG................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CTCTGCTGAGCAGCTCGCTGA..................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GCCCTGGTGGTCTCGCGGA........................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................GACCGCCTTCCCAGAGGCTG.............................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTCTGAGATCCCTAGGACAGC................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GGCTGCTGGAAGTGAGGGG................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................GTGATGGTGTCTCTGAGAT............................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................GTTTATTCTGCAACAGCGAG | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CTTCTCTAGGCCTAAAGCCCTGGTGGTCTCG............................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CTGGAAGTGAAAGTGATGG........................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ............................................................................................................................................................................GCCCTGGTGGTCTCGTGGT........................................................... | 19 | 9 | 0.33 | 0.11 | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | 0.11 | - |
| ............................................................................................................................................................................GCCCTGGTGGTCTCGTGG............................................................ | 18 | 9 | 0.22 | 0.11 | - | - | - | - | - | - | - | - | - | 0.11 | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GCCCTGGTGGTCTCGTTG............................................................ | 18 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - |
| ............................................................................................................................................................................GCCCTGGTGGTCTCGAGGA........................................................... | 19 | 9 | 0.11 | 0.11 | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GCCCTGGTGGTCTCG............................................................... | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| TCATGTGCAAGAAAGACTTTCATCCTGCCTCCAAATCCAATATCAAAAAAGTGAGTAAACAGACCCGGGGAGGGATTAGGCCCACAGGGACCGCCTTCCCAGAGGCTGCTGGAAGTGAAAGTGATGGTGTCTCTGAGATCCCTAGGACAGCCTCCGCTTCTCTAGGCCTAAAGCCCTGGTGGTCTCGCCTTTCTCTGCTGAGCAGCTCGCTGACGCAGCGGGTCCTCAGCGTTTATTCTGCAACAGCGAG ...........................................................(((..((.(((((((....(((((...))).)).)))))))...))...)))..............((..((.....))..))............................................................................................................ ........................................................57...................................................................................142.......................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189782 | TAX577588(Rovira) total RNA. (breast) | SRR037938(GSM510476) 293Red. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191604(GSM715714) 74genomic small RNA (size selected RNA from t. (breast) | SRR040011(GSM532896) G529T. (cervix) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR040009(GSM532894) G727T. (cervix) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR040008(GSM532893) G727N. (cervix) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR553576(SRX182782) source: Testis. (testes) | SRR029132(GSM416761) MB-MDA231. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | TAX577746(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | TAX577739(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | TAX577738(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................GCAGCGGGTCCTCAGCGTTAA............... | 21 | 3.00 | 0.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGAGTAAACAGACCGGC...................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................TTTCATCCTGCCTCCCGA....................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............TATTGGATTTGGAGGCAGGATGAAAGTC................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TTGGATTTGGAGGCAGGATG.................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TTAGGCCCACAGGGAATTG............................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................TGGATTTGGAGGCAGGATGAAA................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................CTGCCTCCAAATCCAATTTGG............................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...................TTGATATTGGATTTGGAGGCAGGATGA............................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GATATTGGATTTGGAGGC.............................................................................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| .........................ATTGGATTTGGAGGCA................................................................................................................................................................................................................. | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |