| (1) AGO2.ip | (5) B-CELL | (5) BRAIN | (4) BREAST | (9) CELL-LINE | (1) CERVIX | (2) HEART | (1) HELA | (4) LIVER | (2) OTHER | (7) SKIN | (1) TESTES |
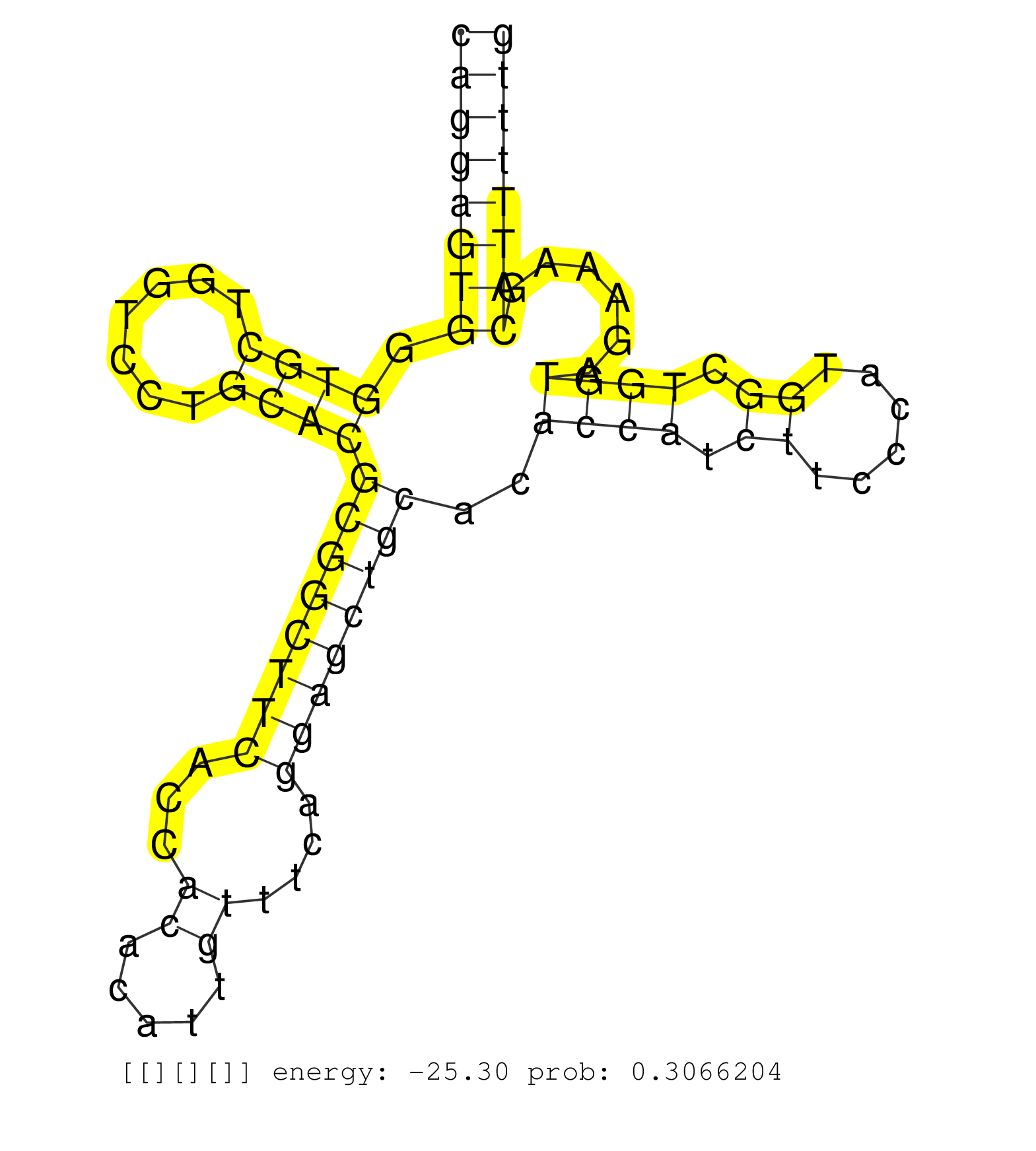
| GGCTTCCTTTGGGTGCAGGGCTGATCCCATAGGTCTTAGCCAGGGAGTGGCTTCCTCAGAGGGAGCTGGGCAGCAGGTGGTGCAGGACTAGAGCCGGGGCAGGAGTGGGTGCTGGTCCTGCACGCGGCTTCACCACACATTGTTTCAGGAGCTGCACACCATCTTCCCATGGCTGGTAGAAAGCATTTTTGGCAGCCTAGATGGTGTCCTCGTTGGCTGGAACCTCCGCTGCTTACAGGGGCGCGTGAAT .....................................................................((....((((((((((((((((.(((...((....)).))).))))))))))))((((((((...((.....))....)))))))).)))).(((..(((....))).)))..)).................................................................. ....................................................................69........................................................................................................................191......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577580(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM532874(GSM532874) G699T. (cervix) | SRR189782 | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR390723(GSM850202) total small RNA. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033728(GSM497073) MALT (MALT413). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................TGGCTGGTAGAAAGCATT............................................................... | 18 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGATGGTGTCCTCGTTGGCTGGAAC........................... | 26 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................ATGGCTGGTAGAAAGCAT................................................................ | 18 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................GTGGGTGCTGGTCCTGCACGCGGCTTCACC.................................................................................................................... | 30 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................ATGGCTGGTAGAAAGCATT............................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CATGGCTGGTAGAAAGCA................................................................. | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CAGGTGGTGCAGGACTAGAGCCG.......................................................................................................................................................... | 23 | 2 | 2.50 | 2.50 | - | - | - | - | - | - | - | - | - | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCTGGGCAGCAGGTGGTGCAGGACCAG............................................................................................................................................................... | 28 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................TGGCTGGTAGAAAGCATTTTTGGCAG....................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CATGGCTGGTAGAAAGCATT............................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................AGAGGGAGCTGGGCAGCAGGTGGTGCAGGACC................................................................................................................................................................. | 32 | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................ATGGCTGGTAGAAAGC.................................................................. | 16 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................AGTGGGTGCTGGTCCTGCACGCGGCTTC....................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TTTTTGGCAGCCTAGATGGTGTCCG........................................ | 25 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................TCCCATGGCTGGTAGAAAGCCTTT.............................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............GTGCAGGGCTGATCCCATAG.......................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CATGGCTGGTAGAAAGCATTTTTGGCA........................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGGCTGGTAGAAAGCATTTTTGG.......................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGGGTGCTGGTCCTGCACGCGGCTTCCCAC................................................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................TGGCAGCCTAGATGGTGTCC......................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................GCACGCGGCTTCACCCGC................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................TGGCTGGTAGAAAGCATTTTT............................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGGTGTCCTCGTTGGCTGGAAC........................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................AGCATTTTTGGCAGCCTAGATGGTGCCCT........................................ | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................TGGGTGCTGGTCCTGCACGCGGCTCC....................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................TGGCTGGTAGAAAGCATTT.............................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CTAGATGGTGTCCTCGTTGGCG................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGGGAGCTGGGCAGCAGGGGTG......................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGATGGTGTCCTCGTTGGCTGGAAC........................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................AGTGGGTGCTGGTCCTGCACGCGGCTCACC..................................................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................AGAGGGAGCTGGGCAGCAGGTGGCG........................................................................................................................................................................ | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................CCCATGGCTGGTAGAAAGCATTTTTGGCA........................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CATGGCTGGTAGAAACCTG................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .............................................................................TGGTGCAGGACTAGAGCCGGGGCAGGAGCGGG............................................................................................................................................. | 32 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................TGGCTGGTAGAAAGCATTTTTTGC......................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CTTTGGGTGCAGGGCTGATCCCATAG.......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CTAGATGGTGTCCTCGTTGGCT................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GCAGGACTAGAGCCGGGGCAGGA.................................................................................................................................................. | 23 | 2 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................CTGCTTACAGGGGCGTTT.... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGGGAGCTGGGCAGCAGGTGGTGCAGGACC................................................................................................................................................................. | 30 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................ACCATCTTCCCATGGGGC........................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................AGTGGGTGCTGGTCCTGCACGCGGCC......................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................AGCAGGTGGTGCAGGTTG................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................TGGGTGCTGGTCCTGCACGCGGCCTC....................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................CATGGCTGGTAGAAAGCATTTTT............................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AGGAGCTGCACACCATCT...................................................................................... | 18 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - |
| ........................................................................................................................................................................................................ATGGTGTCCTCGTTGGCT................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ......................................................CTCAGAGGGAGCTGGGCAGCAGGTG........................................................................................................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CAGCAGGTGGTGCAGGACTAGAGCCGGG........................................................................................................................................................ | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................GGCAGCCTAGATGGTG............................................ | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...........................................................................................................................................................................................................GTGTCCTCGTTGGCTGGAACCTCCGCTGC.................. | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .................................................................................................................................................................................................................TCGTTGGCTGGAACCTCCGCTGCTTACAGGG.......... | 31 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................AGGACTAGAGCCGGGGCAGGA.................................................................................................................................................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TTGTTTCAGGAGCTGCACACCA......................................................................................... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...............................................................................................................................................TTCAGGAGCTGCACACC.......................................................................................... | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CCATCTTCCCATGGCTGGTAGA...................................................................... | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GAGCTGGGCAGCAGGTGGTGCAGG.................................................................................................................................................................... | 24 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................ATGGCTGGTAGAAAG................................................................... | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGCTTCCTTTGGGTGCAGGGCTGATCCCATAGGTCTTAGCCAGGGAGTGGCTTCCTCAGAGGGAGCTGGGCAGCAGGTGGTGCAGGACTAGAGCCGGGGCAGGAGTGGGTGCTGGTCCTGCACGCGGCTTCACCACACATTGTTTCAGGAGCTGCACACCATCTTCCCATGGCTGGTAGAAAGCATTTTTGGCAGCCTAGATGGTGTCCTCGTTGGCTGGAACCTCCGCTGCTTACAGGGGCGCGTGAAT .....................................................................((....((((((((((((((((.(((...((....)).))).))))))))))))((((((((...((.....))....)))))))).)))).(((..(((....))).)))..)).................................................................. ....................................................................69........................................................................................................................191......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577580(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM532874(GSM532874) G699T. (cervix) | SRR189782 | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR390723(GSM850202) total small RNA. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033728(GSM497073) MALT (MALT413). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................CTTCCTCAGAGGGAGCTGA..................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................TGGCTGGAACCTCCGCTC................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................CCTAGATGGTGTCCTAGGG.................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................TTCTACCAGCCATGG..................................................................... | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |