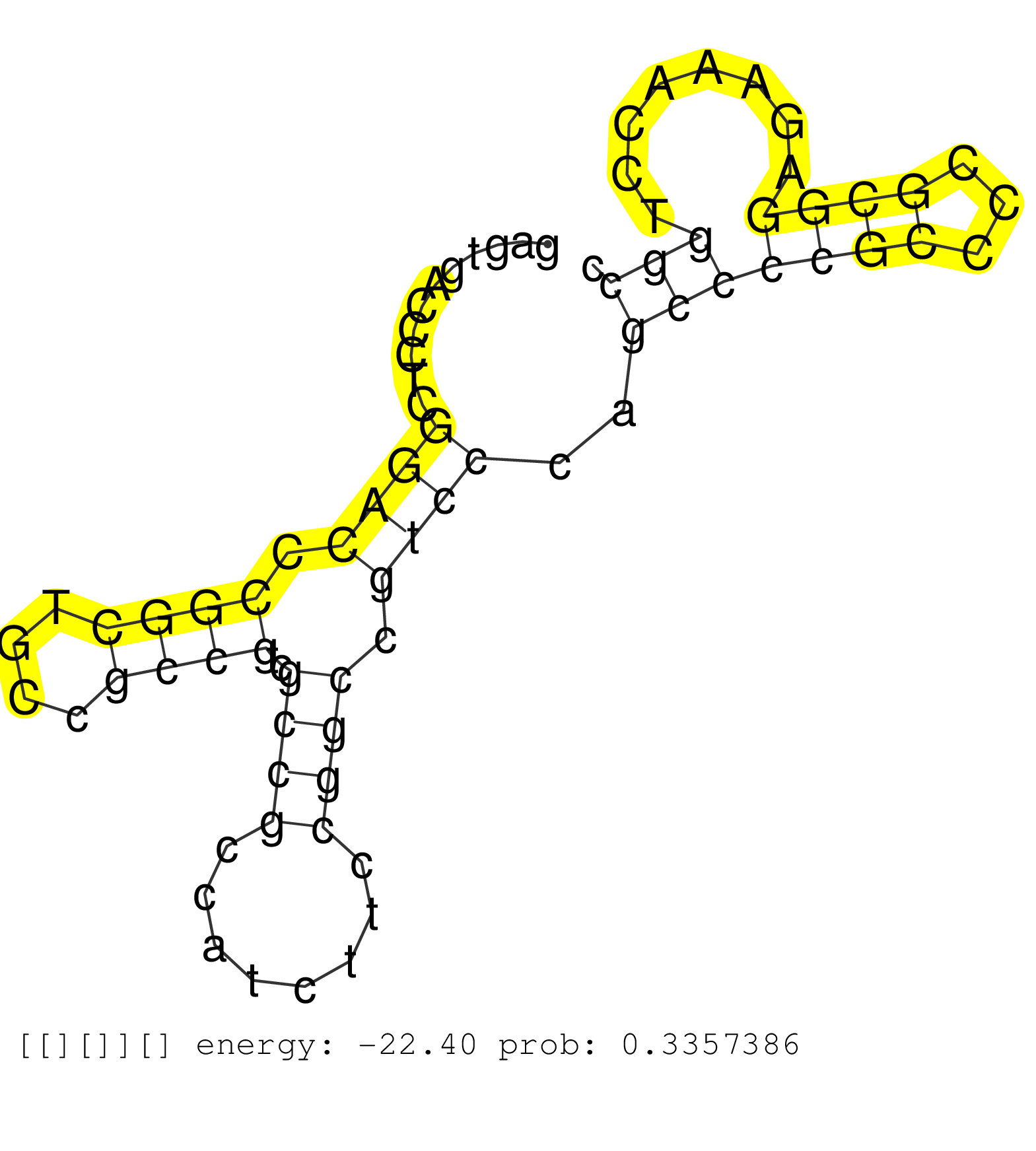
| (2) B-CELL | (2) BREAST | (10) CELL-LINE | (2) FIBROBLAST | (1) LIVER | (1) OTHER | (4) SKIN | (1) TESTES | (1) UTERUS |
| ATCCTCGGAAATGGCAGTAGCTCTCAAAGTAAATAAGTAGCCGCTTGGTAATAAGTTGACTACAATGGGTCCCTTCCATCCTAGAAATGGCAGAAATTTATTTTGACTAGAATGGACACATGGTTTACCTTTTCTGCCTGCAGGGCCTCAGCCAACGCCACTATCTGAGGGCTTACTGAGTGTTTTACCCACTGACATAGGTTCCCACGTAACATCACATCAGACCCAAGGGCCCGTTCATTGCAAGGCG ..................................................................................................................(((((..(((...........)))...(((.((((((.............)))))).))).....))))).................................................................. .............................................................................................94........................................................................................184................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | TAX577589(Rovira) total RNA. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................ATCTGAGGGCTTACTTTT...................................................................... | 18 | 196.00 | 0.00 | 45.00 | 43.00 | 27.00 | 26.00 | 15.00 | 13.00 | 8.00 | 8.00 | 8.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ..................................................................................................................................................................ATCTGAGGGCTTACTTTA...................................................................... | 18 | 6.00 | 0.00 | 1.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................ATCTGAGGGCTTACTTT....................................................................... | 17 | 5.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | |
| ..................................................................................................................................................................ATCTGAGGGCTTACTTTC...................................................................... | 18 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................ATCTGAGGGCTTACTTTCG..................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ................GTAGCTCTCAAAGTAAATAAG..................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TGGACACATGGTTTACCTTTTCTGCC................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................ATCTGAGGGCTTACTTTTA..................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....TCGGAAATGGCAGTAGCTCTG................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..........................AAGTAAATAAGTAGCTA............................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................ATCTGAGGGCTTACTTCT...................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................TATTTTGACTAGAATGG....................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TCCTAGAAATGGCAGA............................................................................................................................................................ | 16 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |
| ATCCTCGGAAATGGCAGTAGCTCTCAAAGTAAATAAGTAGCCGCTTGGTAATAAGTTGACTACAATGGGTCCCTTCCATCCTAGAAATGGCAGAAATTTATTTTGACTAGAATGGACACATGGTTTACCTTTTCTGCCTGCAGGGCCTCAGCCAACGCCACTATCTGAGGGCTTACTGAGTGTTTTACCCACTGACATAGGTTCCCACGTAACATCACATCAGACCCAAGGGCCCGTTCATTGCAAGGCG ..................................................................................................................(((((..(((...........)))...(((.((((((.............)))))).))).....))))).................................................................. .............................................................................................94........................................................................................184................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | TAX577589(Rovira) total RNA. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................AATGGACACATGGTTTACCTTTT..................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ....................................................AAGTTGACTACAATGG...................................................................................................................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .........................................................................................................................................................................................................TCTGATGTGATGTTACGTGGGAA.......................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCCTGCAGGCAGAAAAG......................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ............................................................................................................................................................CCCTCAGATAGTGGC............................................................................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |