| (1) B-CELL | (2) BREAST | (5) CELL-LINE | (1) CERVIX | (1) HEART | (2) HELA | (2) OTHER | (4) SKIN |
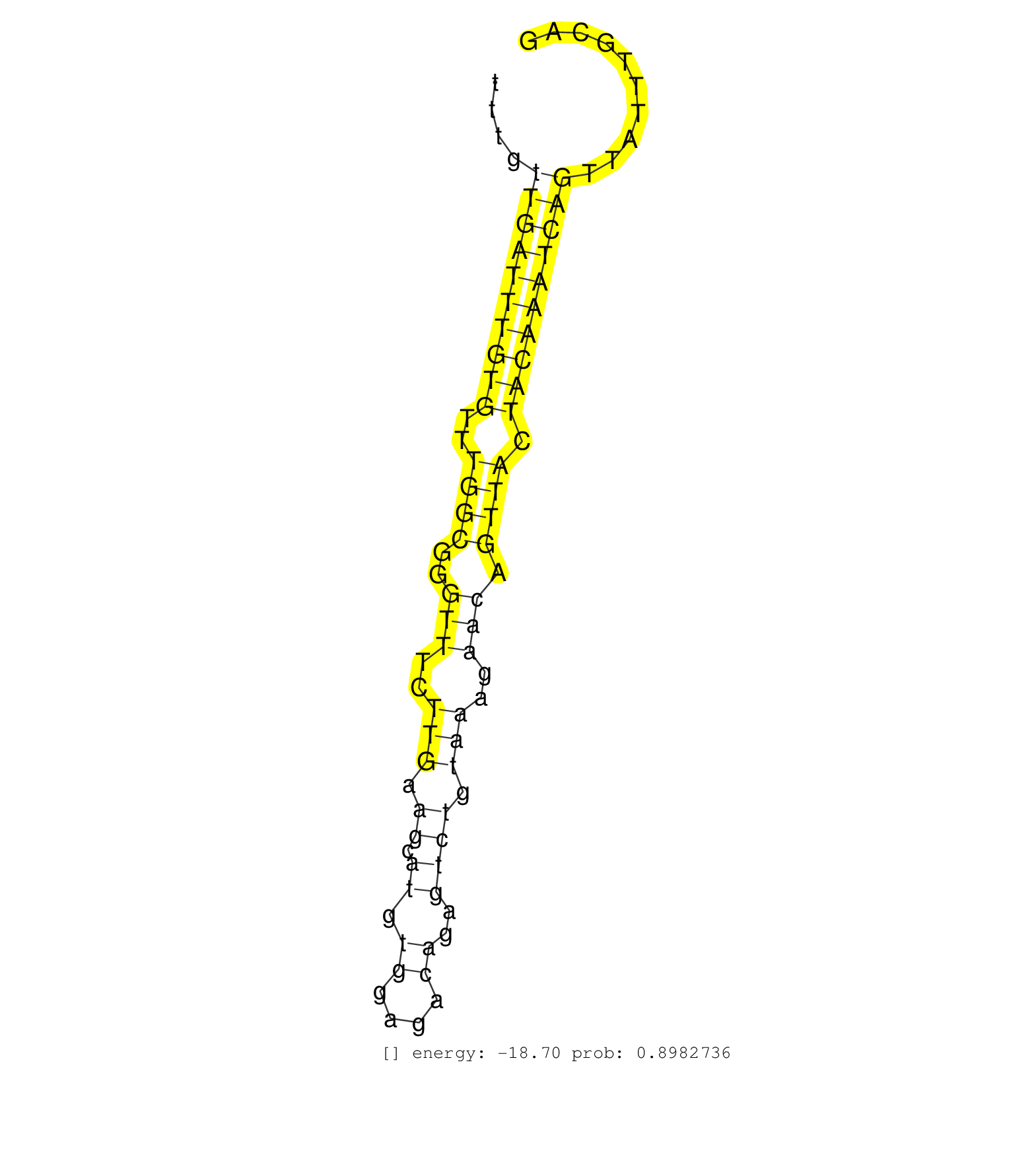
| GCAGAAATAAAATGGTGGATAGTTGGTAGGAGGGGAAAATTAAGGAAAAGTAATATTTGTATACTAAATTTATCAGGTTATGAAAATTTTGCAGAGACATATTTTATTTGTATTGTTGAGGCATAAAATGCTAATTTGTCATTTATTTTGGTCTGTTTCTGAATTATAAATAGTAATAGTTCATTCTGTTTACCTTTCAGATCGTTTGAAAAATGAAAAAGAACCAATGGTGTTGAAACTTAAGGACTGG ........................................................................................................((((((((.....((((((......(((((..((.....))..))))).)))))).....)))))))).............................................................................. .................................................................................................98........................................................................172............................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR343336 | SRR343337 | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189785 | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR189782 | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR343334 | SRR189784 | SRR191436(GSM715546) 173genomic small RNA (size selected RNA from . (breast) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR040028(GSM532913) G026N. (cervix) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................TTTATTTGTATTGTTAGC.................................................................................................................................. | 18 | 39.00 | 0.00 | 27.00 | 10.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................ATTTATTTTGGTCTGTTTCTGAATTATAAATAGTAATAGTTCATTCTGTTTACCTTTCAG.................................................. | 60 | 1 | 7.00 | 7.00 | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TTTATTTGTATTGTTAGCC................................................................................................................................. | 19 | 4.00 | 0.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................GTGTTGAAACTTAAGGACTGG | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTTGGTCTGTTTCTGAATTCTG.................................................................................. | 22 | 3.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TTTATTTTGGTCTGTTTCTGAATTATAAATAGTAATAGTTCATTCTGTTTACCTTTCAG.................................................. | 59 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................AATGGTGTTGAAACTTAAGGACT.. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TTTTATTTGTATTGTTAGTT................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................CTTTCAGATCGTTTGAAAAATGAAA................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............TGGATAGTTGGTAGGA........................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AAAAATGAAAAAGAACCAAT...................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................ACATATTTTATTTGTATTGTTGAGG................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................AAAATTTTGCAGAGACATATTTTAC............................................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................GTTTGAAAAATGAAAAAGAACCAATG..................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................AATGGTGTTGAAACTTAAGGACTGG | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTTTGGTCTGTTTCTGATTTC.................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................................TGTTGAAACTTAAGGACTGG | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CATTCTGTTTACCTTTCAG.................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........ATGGTGGATAGTTGGTAGGAG.......................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| GCAGAAATAAAATGGTGGATAGTTGGTAGGAGGGGAAAATTAAGGAAAAGTAATATTTGTATACTAAATTTATCAGGTTATGAAAATTTTGCAGAGACATATTTTATTTGTATTGTTGAGGCATAAAATGCTAATTTGTCATTTATTTTGGTCTGTTTCTGAATTATAAATAGTAATAGTTCATTCTGTTTACCTTTCAGATCGTTTGAAAAATGAAAAAGAACCAATGGTGTTGAAACTTAAGGACTGG ........................................................................................................((((((((.....((((((......(((((..((.....))..))))).)))))).....)))))))).............................................................................. .................................................................................................98........................................................................172............................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR343336 | SRR343337 | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189785 | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR189782 | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR343334 | SRR189784 | SRR191436(GSM715546) 173genomic small RNA (size selected RNA from . (breast) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR040028(GSM532913) G026N. (cervix) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TTTGGTCTGTTTCTGAAGGCA................................................................................... | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | |
| ...............TGGATAGTTGGTAGGGCG......................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ..........................................................................AGGTTATGAAAATTTTTCTT............................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................AAGGAAAAGTAATATTTTG.............................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| ........................GGTAGGAGGGGAAAATCAG............................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |