| (1) AGO2.ip | (4) B-CELL | (2) BRAIN | (2) BREAST | (21) CELL-LINE | (1) CERVIX | (3) HEART | (1) HELA | (3) LIVER | (1) OTHER | (7) SKIN | (1) XRN.ip |
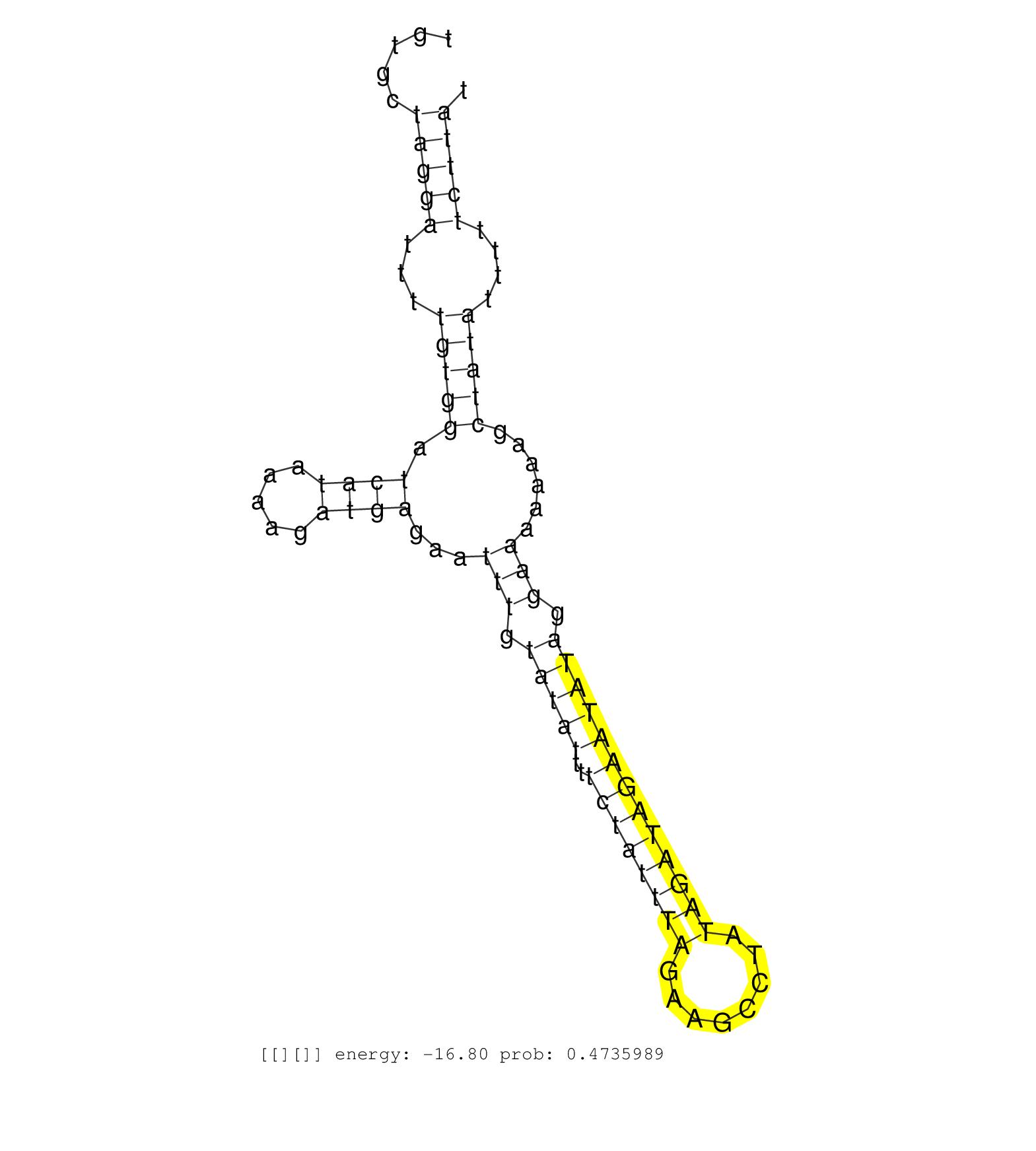
| GTTTGTACAAAGACCACTTTTCCCACTACCTGCAGCCTTTTATCACCCAGGTAAAATTTCACCTCTTTTATAGCAGTTGTGTTTCAGAAACTGATGGAACTGTGCTAGGATTTTGTGGATCATAAAAGATGAGAATTTGTATATTTTCTATTTAGAAGCCTATAGATAGAATATAGGAAAAAAAGCTATATTTTTCTTATAGGCTTCTAAGTAGAAAATGTATTTGCAAGGATATTGTCCAATAGATCTA ........................................................................................................(((((.(((((((...)))))))...((((((......))))))..)))))...((((((.......))))))......................................................................... .................................................................................................98..............................................................................178...................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR189782 | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189786 | SRR189787 | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR343334 | SRR189784 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR040029(GSM532914) G026T. (cervix) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | GSM359192(GSM359192) HepG2_3pM_6. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................TAGAAGCCTATAGATAGAATAT............................................................................ | 22 | 1 | 17.00 | 17.00 | 5.00 | - | - | - | 6.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCTAA........................................ | 22 | 1 | 13.00 | 13.00 | 3.00 | - | 1.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................AGAAGCCTATAGATAGAATAT............................................................................ | 21 | 1 | 9.00 | 9.00 | 6.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCTAT........................................ | 22 | 1 | 9.00 | 3.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................ATTTTTCTTATAGGCTTCTAAG....................................... | 22 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TAGAAGCCTATAGATAGA................................................................................ | 18 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGCTAGGATTTTGTGTTT.................................................................................................................................. | 18 | 4.00 | 0.00 | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................AGAAGCCTATAGATAGAATATA........................................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCTAAT....................................... | 23 | 1 | 3.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCTA......................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TAGAAGCCTATAGATTTTG............................................................................... | 19 | 3.00 | 0.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................TAGAAGCCTATAGATAGAATA............................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCT.......................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCTATC....................................... | 23 | 1 | 2.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................ATTTTTCTTATAGGCTTCCAAG....................................... | 22 | 2.00 | 0.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................ATTTTTCTTATAGGCTTCTAAGA...................................... | 23 | 1 | 2.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCTAAAT...................................... | 24 | 1 | 2.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................ATTTTTCTTATAGGCTTCTATA....................................... | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................ATTTTTCTTATAGGCTTCTAAGT...................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTCTAA......................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...TGTACAAAGACCACTTTTCCCACTAC............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTAA........................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................TAGAAGCCTATAGATAGAATATTTT......................................................................... | 25 | 1 | 1.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................ACTACCTGCAGCCTTTTATCACC........................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCCAAG....................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................ATATTTTTCTTATAGGCTTCTAAT....................................... | 24 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................TATTTTTCTTATAGGCTTCCAA........................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................TAGAAGCCTATAGATTCTG............................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................TTTCCCACTACCTGCAGCCTTTTATCAC............................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ATATTTTCTATTTAGAATAG.......................................................................................... | 20 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................GGAACTGTGCTAGGATTTTGTGGATCAT............................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TTTTTCTTATAGGCTTCTAAG....................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GTACAAAGACCACTTTTCCCACTACCTGCAG....................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ATTTCACCTCTTTTAACAG................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| ........................................................................................................................................................TAGAAGCCTATAGATATTG............................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........GACCACTTTTCCCACGAAT............................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ........................................................................GCAGTTGTGTTTCAGTTT................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ........................................................................................................................................................TAGAAGCCTATAGATATT................................................................................ | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............ACCACTTTTCCCACTACCTG.......................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GACCACTTTTCCCACTAAT............................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................TAGGCTTCTAAGTAGAAAATG.............................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTTTGTACAAAGACCACTTTTCCCACTACCTGCAGCCTTTTATCACCCAGGTAAAATTTCACCTCTTTTATAGCAGTTGTGTTTCAGAAACTGATGGAACTGTGCTAGGATTTTGTGGATCATAAAAGATGAGAATTTGTATATTTTCTATTTAGAAGCCTATAGATAGAATATAGGAAAAAAAGCTATATTTTTCTTATAGGCTTCTAAGTAGAAAATGTATTTGCAAGGATATTGTCCAATAGATCTA ........................................................................................................(((((.(((((((...)))))))...((((((......))))))..)))))...((((((.......))))))......................................................................... .................................................................................................98..............................................................................178...................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR189782 | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189786 | SRR189787 | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR343334 | SRR189784 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR040029(GSM532914) G026T. (cervix) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | GSM359192(GSM359192) HepG2_3pM_6. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................AAAAAAGCTATATTTCAAC..................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................CTACCTGCAGCCTTTGGG............................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................TCCCACTACCTGCAGGAT.................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .........................CTACCTGCAGCCTTTATA............................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |