| (2) AGO2.ip | (19) BREAST | (15) CELL-LINE | (7) CERVIX | (1) HEART | (2) HELA | (6) LIVER | (4) OTHER | (7) SKIN | (1) UTERUS |
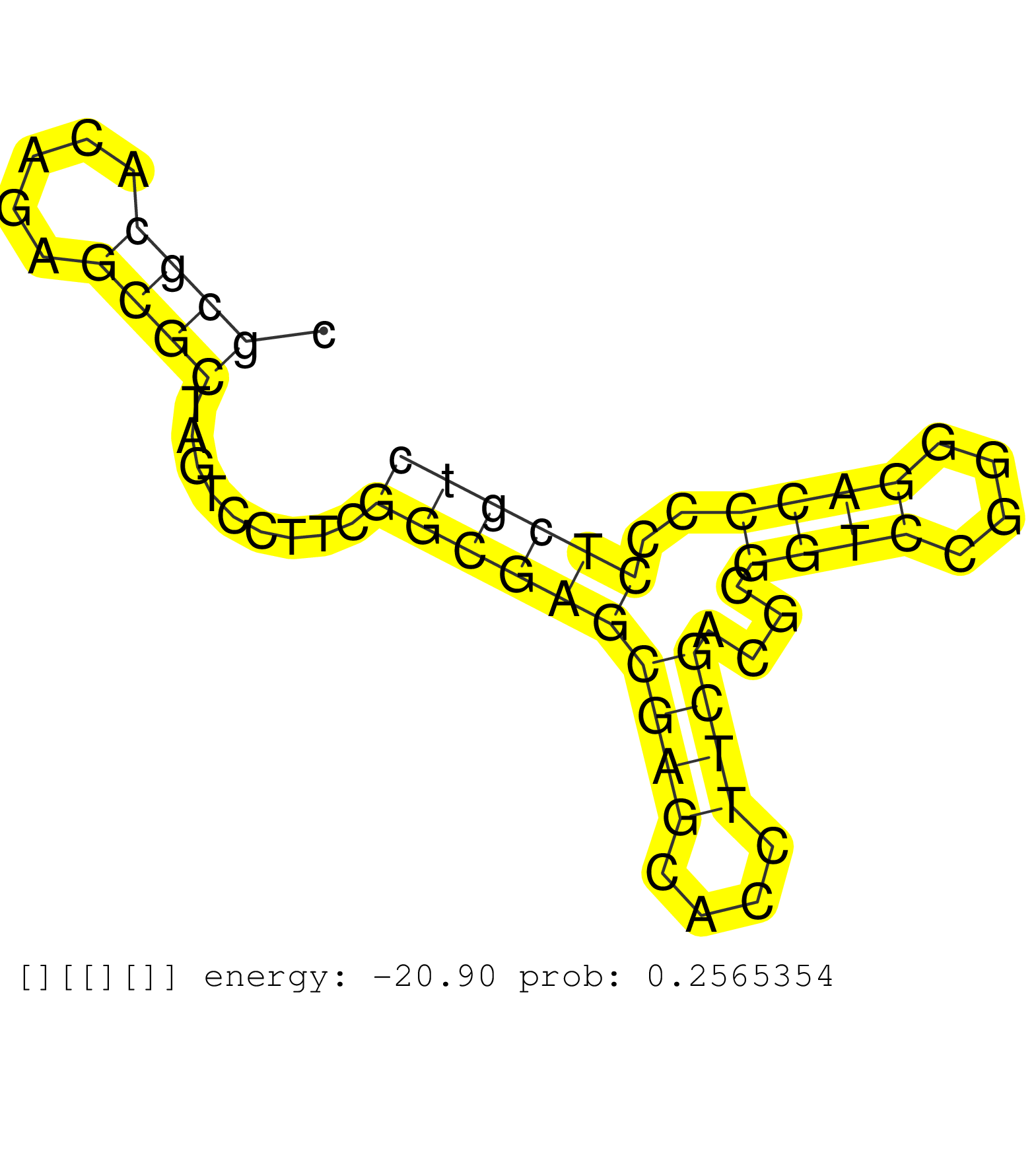
| CTCCCCCAACTCCCGGGCGGTGACTCATCAACGAGCACCAGCGGCCAGAGGTGAGCAGTCCCGGGAAGGGGCCGAGAGGCGGGGCCGCCAGGTCGGGCAGGTGTGCGCTCCGCCCCGCCGCGCGCACAGAGCGCTAGTCCTTCGGCGAGCGAGCACCTTCGACGCGGTCCGGGGACCCCCTCGTCGCTGTCCTCCCGACGCGGACCCGCGTGCCCCAGGCCTCGCGCTGCCCGGCCGGCTCCTCGTGTCC .................................................................................(((((((...(((((..((((((.((((.((((.....((((((.....))))..)).....)))))))).))))))))))))))))))................................................................................ ................................................................................81........................................................................................171............................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM359187(GSM359187) HepG2_2pM_1. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | GSM532876(GSM532876) G547T. (cervix) | GSM359192(GSM359192) HepG2_3pM_6. (cell line) | SRR189785 | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | TAX577742(Rovira) total RNA. (breast) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189784 | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR040029(GSM532914) G026T. (cervix) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | SRR191576(GSM715686) 86genomic small RNA (size selected RNA from t. (breast) | SRR189783 | SRR029129(GSM416758) SW480. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR191500(GSM715610) 7genomic small RNA (size selected RNA from to. (breast) | SRR191606(GSM715716) 84genomic small RNA (size selected RNA from t. (breast) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR040023(GSM532908) G575T. (cervix) | GSM359193(GSM359193) HepG2_3pM_7. (cell line) | SRR444048(SRX128896) Sample 8cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | TAX577741(Rovira) total RNA. (breast) | SRR191620(GSM715730) 167genomic small RNA (size selected RNA from . (breast) | SRR191418(GSM715528) 40genomic small RNA (size selected RNA from t. (breast) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | TAX577579(Rovira) total RNA. (breast) | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR191632(GSM715742) 78genomic small RNA (size selected RNA from t. (breast) | SRR191430(GSM715540) 167genomic small RNA (size selected RNA from . (breast) | GSM359196(GSM359196) m3m7G_ip. (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR040027(GSM532912) G220T. (cervix) | SRR040015(GSM532900) G623T. (cervix) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM532887(GSM532887) G761N. (cervix) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR444041(SRX128889) Sample 2cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR444047(SRX128895) Sample 27_1cDNABarcode: AF-PP-343: ACG CTC TT. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................ACAGAGCGCTAGTCCTTCGGCGAGCGAGCACCTTCG......................................................................................... | 36 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CGAGCACCTTCGACGCGG................................................................................... | 18 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................ACCTTCGACGCGGTCCGG.............................................................................. | 18 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................ACAGAGCGCTAGTCCTTCGGCGAGCGAG................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GCGAGCGAGCACCTTCGA........................................................................................ | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CAGGCCTCGCGCTGCCCGGCCGGC........... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCGGCGAGCGAGCACCTTCGAC....................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ACGCGGTCCGGGGACCCCCT..................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ACGCGGTCCGGGGACCCCCTCGT.................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................CGACGCGGTCCGGGGACCCC....................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TCCTTCGGCGAGCGAGCA............................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................GTCCGGGGACCCCCTCCA.................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................CGCGGTCCGGGGACCCCCTCGTC................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TCCTTCGGCGAGCGAGCACC............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GACGCGGTCCGGGGACCCCCTCGT.................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................ACCTTCGACGCGGTCCGGGG............................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCGGTCCGGGGACCCCCTCGTC................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CGCCAGGTCGGGCAGGTGTG................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CCAGGTCGGGCAGGTGTG................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CCTTCGGCGAGCGAGCACC............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................GTCCCGGGAAGGGGCCGAGAGGCGGGGC..................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCGGTCCGGGGACCCCCTCGT.................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGTCCTTCGGCGAGCGATT................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TCGGCGAGCGAGCACCTTCGA........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GCGAGCACCTTCGACGCGGTCCGG.............................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGGGGCCGAGAGGCGGGGG..................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................ACAGAGCGCTAGTCCTTCGGCGAGCGAGCA............................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................AGAGCGCTAGTCCTTCGGCGAGCGAGCACCTTCGA........................................................................................ | 35 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CGGGGACCCCCTCGT.................................................................. | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CCGGGCGGTGACTCATCAACGAGCACCAGCGG.............................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGGCGGGGCCGCCAG............................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CGCGGTCCGGGGACCCCC...................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CCCGCGTGCCCCAGGCCTCG.......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGGGCCGAGAGGCCTTT...................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................CGCGCTGCCCGGCCGGCTCCTCGT.... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TCCCGGGCGGTGACTTCA.............................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................TCCCGACGCGGACCCGCG........................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ACGCGGTCCGGGGACCCCCTCGTC................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CGCGGTCCGGGGACCCCCTCGT.................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................CGGGAAGGGGCCGAGAGGCGGGA...................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................CGACGCGGTCCGGGGAC.......................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TCCGGGGACCCCCTCGAATT............................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................ACGCGGTCCGGGGACCCCCTC.................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGTCCTTCGGCGAGCGAG................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CCTTCGGCGAGCGAGCACCT............................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................CCGCCCCGCCGCGCGTGCG.......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................GCCCCGCCGCGCGCA............................................................................................................................ | 15 | 7 | 0.29 | 0.29 | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| ..............................................................................................................CGCCCCGCCGCGCGCA............................................................................................................................ | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GCCCCGCCGCGCGCAAAG......................................................................................................................... | 18 | 7 | 0.14 | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - |
| CTCCCCCAACTCCCGGGCGGTGACTCATCAACGAGCACCAGCGGCCAGAGGTGAGCAGTCCCGGGAAGGGGCCGAGAGGCGGGGCCGCCAGGTCGGGCAGGTGTGCGCTCCGCCCCGCCGCGCGCACAGAGCGCTAGTCCTTCGGCGAGCGAGCACCTTCGACGCGGTCCGGGGACCCCCTCGTCGCTGTCCTCCCGACGCGGACCCGCGTGCCCCAGGCCTCGCGCTGCCCGGCCGGCTCCTCGTGTCC .................................................................................(((((((...(((((..((((((.((((.((((.....((((((.....))))..)).....)))))))).))))))))))))))))))................................................................................ ................................................................................81........................................................................................171............................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM359187(GSM359187) HepG2_2pM_1. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | GSM532876(GSM532876) G547T. (cervix) | GSM359192(GSM359192) HepG2_3pM_6. (cell line) | SRR189785 | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | TAX577742(Rovira) total RNA. (breast) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189784 | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR040029(GSM532914) G026T. (cervix) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | SRR191576(GSM715686) 86genomic small RNA (size selected RNA from t. (breast) | SRR189783 | SRR029129(GSM416758) SW480. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR191500(GSM715610) 7genomic small RNA (size selected RNA from to. (breast) | SRR191606(GSM715716) 84genomic small RNA (size selected RNA from t. (breast) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR040023(GSM532908) G575T. (cervix) | GSM359193(GSM359193) HepG2_3pM_7. (cell line) | SRR444048(SRX128896) Sample 8cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | TAX577741(Rovira) total RNA. (breast) | SRR191620(GSM715730) 167genomic small RNA (size selected RNA from . (breast) | SRR191418(GSM715528) 40genomic small RNA (size selected RNA from t. (breast) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | TAX577579(Rovira) total RNA. (breast) | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR191632(GSM715742) 78genomic small RNA (size selected RNA from t. (breast) | SRR191430(GSM715540) 167genomic small RNA (size selected RNA from . (breast) | GSM359196(GSM359196) m3m7G_ip. (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR040027(GSM532912) G220T. (cervix) | SRR040015(GSM532900) G623T. (cervix) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM532887(GSM532887) G761N. (cervix) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR444041(SRX128889) Sample 2cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR444047(SRX128895) Sample 27_1cDNABarcode: AF-PP-343: ACG CTC TT. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................AGCGCGAGGCCTGGGGC...................... | 17 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GCTCGCTCGCCGAAGGAC................................................................................................ | 18 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................CCGACGCGGACCCGCGTGCCCCAGGCCTAG.......................... | 30 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .ACCGCCCGGGAGTTGGGGGA..................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CGCCCCGCCGCGCGCAGCG......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................GAAGGGGCCGAGAGGG.......................................................................................................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................GTCCTTCGGCGAGCGAAG................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................CGCGCGGCGGGGCGG.............................................................................................................................. | 15 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CTCCGCCCCGCCGCGAT.............................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................GACCCGCGTGCCCCATT............................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................CTCGCGCTGCCCGGCCAAT........... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................GGGTCCGCGTCGGGAGG........................................... | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................GCTCCGCCCCGCCGCACCA............................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................GTCGCTGTCCTCCCGTGA.................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................CGCGGACCCGCGTGCCCC.................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........TGAGTCACCGCCCGGGAG............................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TGCCCGGCCGGCTCCTCCTC... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................CTTCCCGGGACTGCTCACCT...................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CCGGGCGGTGACTCACCAG........................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................CGGCGGGGCGGAGCGC.................................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CTCCGCCCCGCCGCGCGGGG........................................................................................................................... | 20 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | |
| ..............................................................................................................................................................................................GTCCGCGTCGGGAGG............................................. | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGCGCGCGGCGGGGCGG............................................................................................................................ | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - |
| ...........................................................................................................CTCCGCCCCGCCGCGCGGGGT.......................................................................................................................... | 21 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................CGCGCGGCGGGGCGGAG.............................................................................................................................. | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - |
| ..............................................................................................................................................................................................................................CGGCCGGGCAGCGCG............. | 15 | 6 | 0.17 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GCGGCGGGGCGGAGCG................................................................................................................................. | 16 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |