| (3) BREAST | (12) CELL-LINE | (1) CERVIX | (1) HEART | (1) HELA | (1) LIVER | (2) OTHER | (1) RRP40.ip | (3) SKIN | (1) XRN.ip |
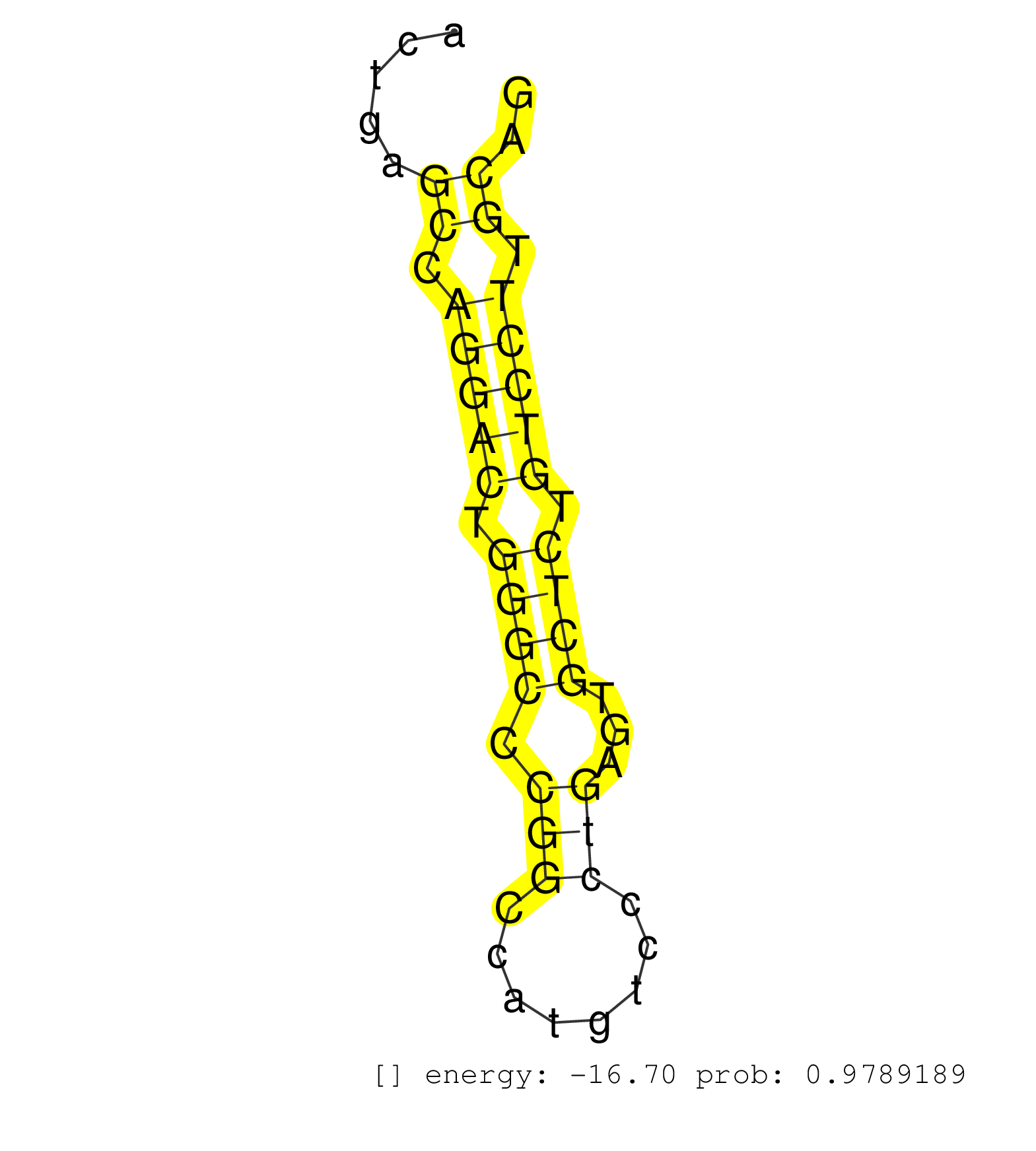
| AGCAGACTCACCCTGGGTCTGTGAGGGAAGCCTCCTCAGATGCGCCAAGCCCTGGGAGGTTGGGGAGGAGGCGGGTCTCTGCCCACTGGGCTCCCAGCCAAACACCTCATCTCCTAGGAAGGCTAGAGATGACCTCGGGCCTCAGGCTGACTGAGCCAGGACTGGGCCCGGCCATGTCCCTGAGTGCTCTGTCCTTGCAGGTTTGGGGACGACATCCCTGGAATGGAAGGCCTGGGAACAGGTATGACGG ..........................................................................................................................................................((.(((((.((((.(((........)))...)))).))))).)).................................................... .....................................................................................................................................................150...............................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189784 | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | TAX577453(Rovira) total RNA. (breast) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR040035(GSM532920) G001T. (cervix) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................GAGTGCTCTGTCCTTGCAGG................................................. | 20 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCCAGGACTGGGCCCGGC.............................................................................. | 18 | 1 | 7.00 | 7.00 | 5.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GACTGAGCCAGGACTGGGCC.................................................................................. | 20 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCAGGCTGACTGAGCCAGGA......................................................................................... | 20 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCCAGGACTGGGCCCGGCC............................................................................. | 19 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GAGCCAGGACTGGGCCCGGC.............................................................................. | 20 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................AGTGCTCTGTCCTTGCAGGT................................................ | 20 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CAGGACTGGGCCCGGCCAT........................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................AGTGCTCTGTCCTTGCAGGTGATG............................................ | 24 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GACTGAGCCAGGACTGG..................................................................................... | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GACCTCGGGCCTCAGGC....................................................................................................... | 17 | 3 | 1.33 | 1.33 | - | - | - | - | - | - | - | - | 1.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAGGCTGACTGAGCCAGGACTGGGCCCGGC.............................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCCAGGACTGGGCCCGG............................................................................... | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AGGCTGACTGAGCCAGGACTGG..................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCCAGGACTGGGCCCTGCC............................................................................. | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................AGGACTGGGCCCGGCCGG........................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................CTGTCCTTGCAGGTTTGGG........................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................CTGACTGAGCCAGGACTGG..................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................GGGCCTCAGGCTGACACCG............................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................CTGACTGAGCCAGGACTG...................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................CCCGGCCATGTCCCTGAGTG................................................................ | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CCTAGGAAGGCTAGAGA......................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TCCCTGAGTGCTCTGTCCTTGCAGGTTTG............................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TCGGGCCTCAGGCTGGGCA................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................AGGACTGGGCCCGGCCATG.......................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GGGCCTCAGGCTGACTGAGCCAGGACTGG..................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................ACCTCGGGCCTCAGGC....................................................................................................... | 16 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ..............................................................................................................................AGATGACCTCGGGCCGAT.......................................................................................................... | 18 | 5 | 0.60 | 0.20 | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | 0.20 |
| ..............................................................................................................................AGATGACCTCGGGCCGATC......................................................................................................... | 19 | 5 | 0.60 | 0.20 | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - | - | - |
| .............................................................................................................................................................................ATGTCCCTGAGTGCTCTG........................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................AGATGACCTCGGGCC............................................................................................................. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| AGCAGACTCACCCTGGGTCTGTGAGGGAAGCCTCCTCAGATGCGCCAAGCCCTGGGAGGTTGGGGAGGAGGCGGGTCTCTGCCCACTGGGCTCCCAGCCAAACACCTCATCTCCTAGGAAGGCTAGAGATGACCTCGGGCCTCAGGCTGACTGAGCCAGGACTGGGCCCGGCCATGTCCCTGAGTGCTCTGTCCTTGCAGGTTTGGGGACGACATCCCTGGAATGGAAGGCCTGGGAACAGGTATGACGG ..........................................................................................................................................................((.(((((.((((.(((........)))...)))).))))).)).................................................... .....................................................................................................................................................150...............................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189784 | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | TAX577453(Rovira) total RNA. (breast) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR040035(GSM532920) G001T. (cervix) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................GGGGAGGAGGCGGGTCTCAG......................................................................................................................................................................... | 20 | 4.00 | 0.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................GCTTGGCGCATCTGAGGA........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................GAGTGCTCTGTCCTTGTG................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................CCTCCTCAGATGCGCCAAGCCCTGGGA................................................................................................................................................................................................. | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................GTGCTCTGTCCTTGCGACA................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .................................................................................................CCAAACACCTCATCTCCGGGG.................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |