| (5) B-CELL | (4) BREAST | (20) CELL-LINE | (2) CERVIX | (2) HELA | (4) LIVER | (1) OTHER | (10) SKIN | (1) XRN.ip |
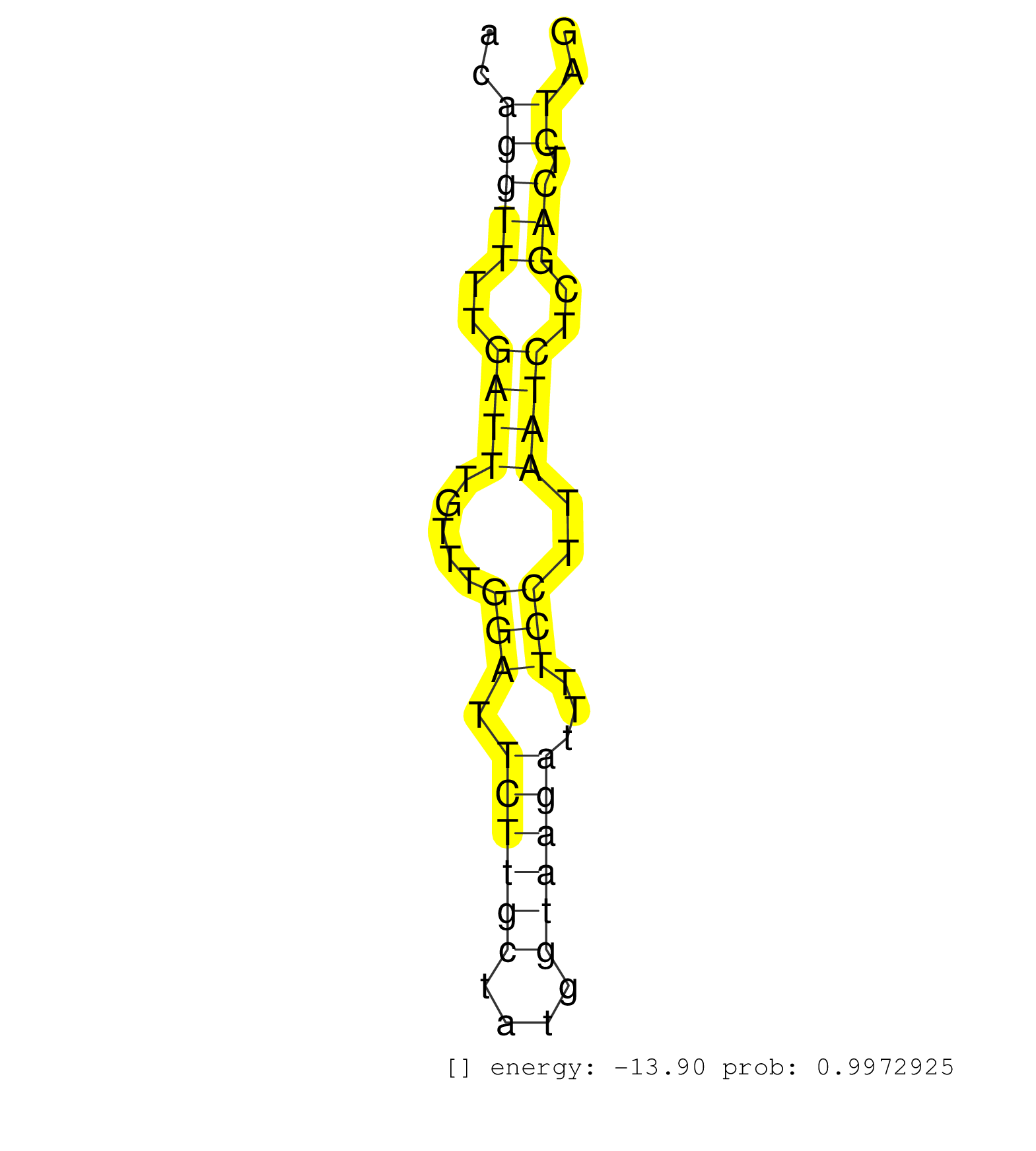
| GGTCTTGAACTCCTGACCTCAGGTGATACACCCGCCTCGGCCTCCCACAGTGCTGGGATTACAGGCATGAGCCACCGTACCTGGCTATATTTCACAAATGTTTAATGTGTTGTGGTTTGACACAGATATAGACTGATGGTACAGGTTTTGATTTGTTTGGATTCTTGCTATGGTAAGATTTTCCTTAATCTCGACTCTAGTTCTTCAGATTATTAAGGAATCCCAGCAGCAGCATGGTTTACGGCATGGA ...............................................................(((....)))...(((((.(((((((((.((((.........))))..(((......))).))))))).))...))))).((((..((((.....(((.((((((....))))))...)))..))))..))))...................................................... .......................................................56..............................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189786 | SRR029125(GSM416754) U2OS. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR189787 | SRR037937(GSM510475) 293cand2. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191576(GSM715686) 86genomic small RNA (size selected RNA from t. (breast) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR040023(GSM532908) G575T. (cervix) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR189785 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR038859(GSM458542) MM386. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | DRR001483(DRX001037) "Hela long cytoplasmic cell fraction, control. (hela) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR040024(GSM532909) G613N. (cervix) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR189782 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TTTCCTTAATCTCGACTCTAG.................................................. | 21 | 1 | 19.00 | 19.00 | - | - | 5.00 | - | - | 4.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................TTCCTTAATCTCGACTCTAG.................................................. | 20 | 1 | 12.00 | 12.00 | - | - | 1.00 | 1.00 | - | - | 2.00 | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTCCTTAATCTCGACTCTAGA................................................. | 22 | 1 | 5.00 | 19.00 | - | - | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTTAATCTCGACTCTAGTTCTTCAG.......................................... | 28 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CAGCAGCAGCATGGTTTACA....... | 20 | 4.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................ATGGTACAGGTTTTGATTTGTTTGGAC........................................................................................ | 27 | 4.00 | 0.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................ACACCCGCCTCGGCCTCCCACAGTGCTGGGAT............................................................................................................................................................................................... | 32 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ATCCCAGCAGCAGCATGGTT........... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTTTCCTTAATCTCGACTCTAG.................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTCCTTAATCTCGACTCTAGTTTT.............................................. | 25 | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................AATCCCAGCAGCAGCATGGTTTACGGCACGG. | 31 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................AGCAGCAGCATGGTTTACA....... | 19 | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TTCCTTAATCTCGACTCTAGA................................................. | 21 | 1 | 2.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................................GCAGCAGCATGGTTTACGGCA.... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTCCTTAATCTCGACTCTA................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTTAATCTCGACTCT.................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACAGGCATGAGCCACCGTACCTGGTG.................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................CCCACAGTGCTGGGATTACAGGATA...................................................................................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................TCGGCCTCCCACAGTGCTCTG................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................TTAAGGAATCCCAGCAGCAG.................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................AGGAATCCCAGCAGCAGCACGG............. | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................AGGTTTTGATTTGTTTGGATTCTTGCTATGGTAAGATTTTCCTTAATCTCGACTCTAG.................................................. | 58 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACAGGCATGAGCCACCGTACCGGTC..................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................TACACCCGCCTCGGCCCCC............................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....TTGAACTCCTGACCTCAGGG.................................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................CAGCAGCAGCATGGTTACA........ | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................GATTTTCCTTAATCTCGACTCTAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................................TTCCTTAATCTCGACTCTA................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CAGCAGCAGCATGGTTCAC........ | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................TCGACTCTAGTTCTTCAG.......................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CCCGCCTCGGCCTCCCACAGTGCTT................................................................................................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................AGGTTTTGATTTGTTTGGATTCTT.................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTCCTTAATCTCGACTCTAGTTCTTCAG.......................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................AGGAATCCCAGCAGCAGCATGGCTC.......... | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................CCCGCCTCGGCCTCCCACAGTGCTGGGGC............................................................................................................................................................................................... | 29 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................ATGGTACAGGTTTTGATTTGTCCGG.......................................................................................... | 25 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................CGGCCTCCCACAGTGCTGGGATTACAGG......................................................................................................................................................................................... | 28 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTTTGATTTGTTTGGATTTA..................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................ATTATTAAGGAATCCCAGCAG..................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CAGTGCTGGGATTACAGGCATGAGAGGG............................................................................................................................................................................... | 28 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................ACCCGCCTCGGCCTCCCACAGTGCGG................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TTCCTTAATCTCGACTCTAGAAA............................................... | 23 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................CAGCAGCATGGTTTATC....... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................ACAGATATAGACTGATGGTACA........................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TAAGGAATCCCAGCAGCAGCATGGTTTACGG...... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GTACCTGGCTATATT............................................................................................................................................................... | 15 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - |
| ..................................................................................................................................................................................................................................CAGCAGCATGGTTTAC........ | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ............................................................ACAGGCATGAGCCACCGTACCTGGCTA................................................................................................................................................................... | 27 | 6 | 0.17 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGTCTTGAACTCCTGACCTCAGGTGATACACCCGCCTCGGCCTCCCACAGTGCTGGGATTACAGGCATGAGCCACCGTACCTGGCTATATTTCACAAATGTTTAATGTGTTGTGGTTTGACACAGATATAGACTGATGGTACAGGTTTTGATTTGTTTGGATTCTTGCTATGGTAAGATTTTCCTTAATCTCGACTCTAGTTCTTCAGATTATTAAGGAATCCCAGCAGCAGCATGGTTTACGGCATGGA ...............................................................(((....)))...(((((.(((((((((.((((.........))))..(((......))).))))))).))...))))).((((..((((.....(((.((((((....))))))...)))..))))..))))...................................................... .......................................................56..............................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189786 | SRR029125(GSM416754) U2OS. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR189787 | SRR037937(GSM510475) 293cand2. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191576(GSM715686) 86genomic small RNA (size selected RNA from t. (breast) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR040023(GSM532908) G575T. (cervix) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR189785 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR038859(GSM458542) MM386. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | DRR001483(DRX001037) "Hela long cytoplasmic cell fraction, control. (hela) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR040024(GSM532909) G613N. (cervix) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR189782 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TACAGGCATGAGCCACCGTACCGA....................................................................................................................................................................... | 24 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................AGCACTGTGGGAGGCCG.................................................................................................................................................................................................... | 17 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGCCTGTAATCCCAGCACTGTGGGAGG...................................................................................................................................................................................... | 28 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................ACCCGCCTCGGCCTCCCATTTC....................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........ACTCCTGACCTCAGGTGATACCC........................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........CCTGACCTCAGGTGATACACCCGCCTTGC.................................................................................................................................................................................................................. | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................GCACTGTGGGAGGCCGAGGC..................................................................................................................................................................................................... | 20 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................ACCCGCCTCGGCCTCCCGGG......................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ..............GACCTCAGGTGATACACCGAGG...................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ..................................CCCAGCACTGTGGGAGGCCGAGG................................................................................................................................................................................................. | 23 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CCCGCCTCGGCCTCCCACAGTGAG.................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................AGCAGCAGCATGGTTTATTCT..... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................CCGCCTCGGCCTCCCACCTGG...................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |