| (2) B-CELL | (4) BREAST | (38) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (2) HEART | (11) HELA | (6) LIVER | (1) RRP40.ip | (6) SKIN | (1) TESTES | (1) XRN.ip |
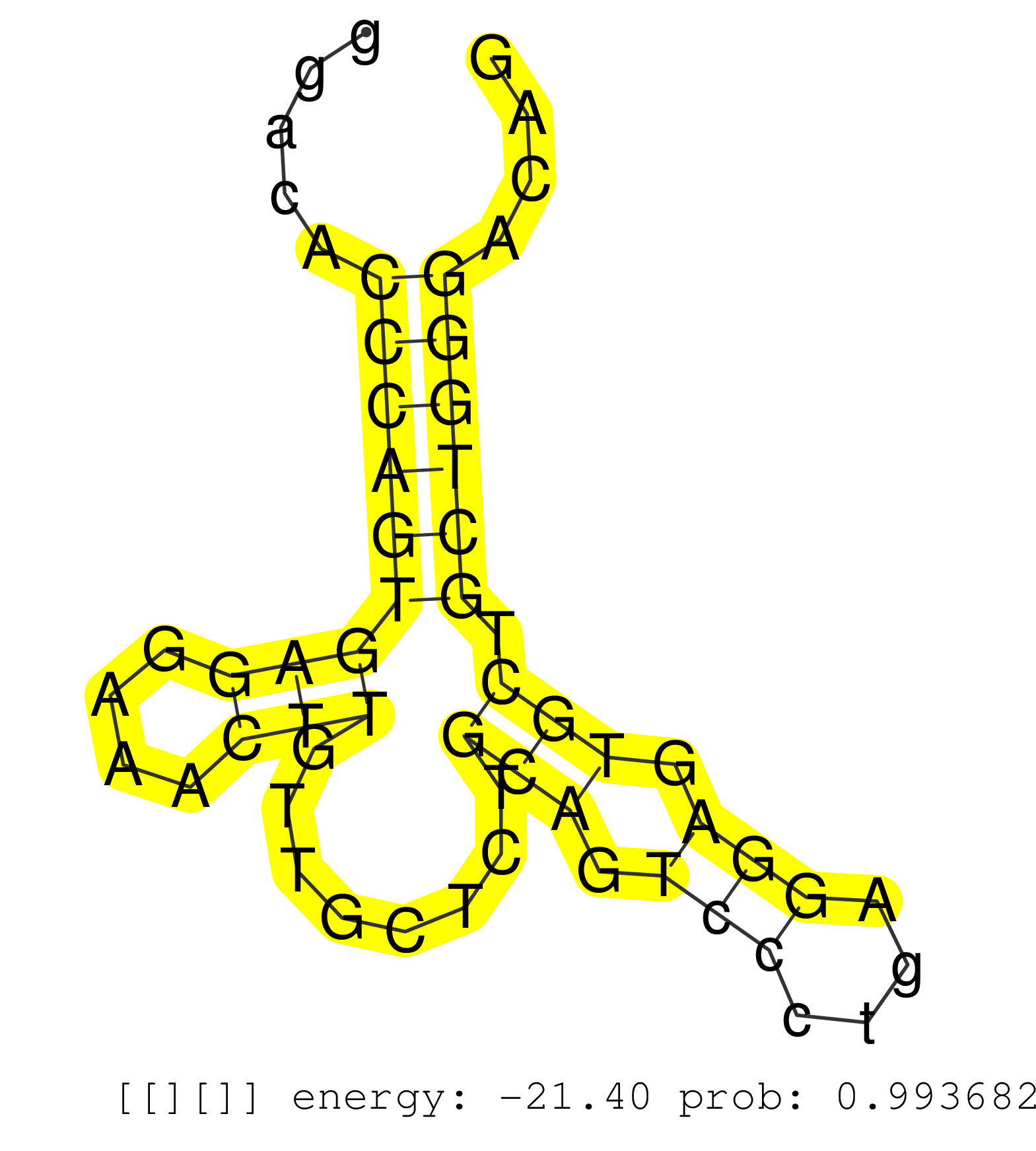
| TCTGGATGTTGTCCTGGTGCCCATCATTCATAGGGAGAGGTCCGTCCCAGGGACACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGAGTGCTGCTGGGACAGTGGTTCTCTCGAGATAGCTCCTATATGGAAATACCCTCGATTTGCCAAGTTAGAGCCTTTAGCCGGTCACATGTGCTTTCTGTCCCTCCCAGGTTTGGAAACGATGTGCAGCACTTCAAGGTGCTCCGAGATGGAGCCGGGA .......................................................(((((((((....)))........(((.(((....))).))).)))))).................................................................................................................................................. ..................................................51.......................................................108............................................................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM359192(GSM359192) HepG2_3pM_6. (cell line) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | GSM359189(GSM359189) HepG2_2pm_3. (cell line) | GSM359190(GSM359190) HepG2_3pM_4. (cell line) | GSM359193(GSM359193) HepG2_3pM_7. (cell line) | GSM359204(GSM359204) HepG2_nucl_low. (cell line) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | GSM359188(GSM359188) HepG2_2pM_2. (cell line) | GSM359187(GSM359187) HepG2_2pM_1. (cell line) | GSM359207(GSM359207) HepG2_tot_up. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | DRR001487(DRX001041) "Hela long nuclear cell fraction, LNA(+)". (hela) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | GSM359205(GSM359205) HepG2_nucl_up. (cell line) | GSM359200(GSM359200) hepg2_tap_pasr_a. (cell line) | GSM359201(GSM359201) hepg2_tap_pasr_b. (cell line) | GSM359206(GSM359206) HepG2_tot_low. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359173(GSM359173) hela_10_1. (hela) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | GSM359209(GSM359209) hepg2_biotin_el_hl_a. (cell line) | GSM359175(GSM359175) hela_5_pct. (hela) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | GSM359177(GSM359177) hela_nucl_a. (hela) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR037936(GSM510474) 293cand1. (cell line) | GSM359210(GSM359210) hepg2_biotin_el_hl_b. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR040011(GSM532896) G529T. (cervix) | SRR029129(GSM416758) SW480. (cell line) | DRR001484(DRX001038) "Hela long nuclear cell fraction, control". (hela) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM532874(GSM532874) G699T. (cervix) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | GSM359186(GSM359186) hepg2_untrtd. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR029127(GSM416756) A549. (cell line) | GSM359185(GSM359185) hepg2_tap_pasr. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR029130(GSM416759) DLD2. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577589(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | GSM359181(GSM359181) hela_phil_smrna_nuc. (hela) | GSM359195(GSM359195) HepG2_TAP_depl. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGT...................................................................................................................................................................... | 30 | 1 | 115.00 | 115.00 | - | 17.00 | 16.00 | - | 12.00 | 14.00 | 16.00 | 1.00 | 11.00 | 6.00 | 6.00 | - | - | 2.00 | - | - | - | 5.00 | - | - | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGA............................................................................................................................................................. | 39 | 1 | 33.00 | 33.00 | 32.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCT............................................................................................................................................................................. | 23 | 1 | 31.00 | 31.00 | - | 4.00 | 2.00 | 1.00 | 5.00 | 3.00 | - | - | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................CCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAG............................................................................................................................................................... | 36 | 1 | 24.00 | 24.00 | - | - | - | 1.00 | - | - | - | 11.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | 3.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCC.................................................................................................................................................................... | 32 | 1 | 17.00 | 17.00 | - | - | - | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGT................................................................................................................................................................................. | 19 | 1 | 10.00 | 10.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGAG............................................................................................................................................................ | 40 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 5.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTG.......................................................................................................................................................................... | 26 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCA........................................................................................................................................................................ | 28 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TTGTTGCTCTGCAGTCCCTGAGGAG............................................................................................................................................................ | 25 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CCCAGTGAGGAAACTTGTTGCT............................................................................................................................................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAG....................................................................................................................................................................... | 29 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAACTTGTTGCTCTGCAGTCCCTGAGGAG............................................................................................................................................................ | 29 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGG.............................................................................................................................................................. | 38 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................CACCCAGTGAGGAAACTTGTTGCT............................................................................................................................................................................. | 24 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................CCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGG.............................................................................................................................................................. | 36 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GGAAACGATGTGCAGCACT........................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AGGAGTGCTGCTGGGACAG.............................................................................................................................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AGGAAACTTGTTGCTCTGCAGTCCCTGAGGAG............................................................................................................................................................ | 32 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGC......................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTTTGGAAACGATGTGCAG............................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GAAACGATGTGCAGCACTTCAA....................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GTTGCTCTGCAGTCCCTGAGGAG............................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGAGAA.......................................................................................................................................................... | 38 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................CACCCAGTGAGGAAACTTGTTGCTCTGCAGTCC.................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CTTGTTGCTCTGCAGTCC.................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TTGGAAACGATGTGCAGCA............................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGAGA........................................................................................................................................................... | 41 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GAGGAAACTTGTTGCTCT........................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCC................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................CGAGATGGAGCCGGGCGGC | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................AAACTTGTTGCTCTGCAGTCCCTGAGGA............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTTGGAAACGATGTGCAGCA............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CTGGGACAGTGGTTCTCTCGAGAT............................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GAGGAAACTTGTTGCTCTGCAGTCCCTGAGGA............................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTT................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TTGTTGCTCTGCAGTCCCTGAGGAGA........................................................................................................................................................... | 26 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TTGGAAACGATGTGC................................. | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGC...................................................................................................................................................................... | 30 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTTTGGAAACGATGTGC................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGCAGTCCCTGAGGAACC.......................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGTGGA............................................................................................................................................................. | 39 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................GGAAACTTGTTGCTCTGCAGTCCCTGAGG.............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGAAACGATGTGCAGCACTTT......................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .....................................................................TTGTTGCTCTGCAGTCCCTGAGGAGTGCTGC...................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................TGCTCTGCAGTCCCTGAGGA............................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AACGATGTGCAGCACTTCAAGGTG................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGA............................................................................................................................................................. | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................AATACCCTCGATTTGCTTGT............................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ...........................................................................................................................................................................................................TGGAAACGATGTGCAGCACTTCAAGGT.................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................CGGTCACATGTGCTTTCTGTCCCTCCCAGG................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGT............................................................................................................................................................. | 39 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TAGCCGGTCACATGTGCTTTCTGTCCCTCCCAG.................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CCCAGTGAGGAAACTTGTTGCTCTGCAGT...................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGAG............................................................................................................................................................ | 39 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGA............................................................................................................................................................. | 38 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ACGATGTGCAGCACTTCAA....................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................ACTTGTTGCTCTGCAGTCCCTGAGG.............................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................AAGGTGCTCCGAGAT.......... | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GGTGCTCCGAGATGG........ | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| ......................................................ACCCAGTGAGGAAAC..................................................................................................................................................................................... | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| TCTGGATGTTGTCCTGGTGCCCATCATTCATAGGGAGAGGTCCGTCCCAGGGACACCCAGTGAGGAAACTTGTTGCTCTGCAGTCCCTGAGGAGTGCTGCTGGGACAGTGGTTCTCTCGAGATAGCTCCTATATGGAAATACCCTCGATTTGCCAAGTTAGAGCCTTTAGCCGGTCACATGTGCTTTCTGTCCCTCCCAGGTTTGGAAACGATGTGCAGCACTTCAAGGTGCTCCGAGATGGAGCCGGGA .......................................................(((((((((....)))........(((.(((....))).))).)))))).................................................................................................................................................. ..................................................51.......................................................108............................................................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM359192(GSM359192) HepG2_3pM_6. (cell line) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | GSM359189(GSM359189) HepG2_2pm_3. (cell line) | GSM359190(GSM359190) HepG2_3pM_4. (cell line) | GSM359193(GSM359193) HepG2_3pM_7. (cell line) | GSM359204(GSM359204) HepG2_nucl_low. (cell line) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | GSM359188(GSM359188) HepG2_2pM_2. (cell line) | GSM359187(GSM359187) HepG2_2pM_1. (cell line) | GSM359207(GSM359207) HepG2_tot_up. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | DRR001487(DRX001041) "Hela long nuclear cell fraction, LNA(+)". (hela) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | GSM359205(GSM359205) HepG2_nucl_up. (cell line) | GSM359200(GSM359200) hepg2_tap_pasr_a. (cell line) | GSM359201(GSM359201) hepg2_tap_pasr_b. (cell line) | GSM359206(GSM359206) HepG2_tot_low. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359173(GSM359173) hela_10_1. (hela) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | GSM359209(GSM359209) hepg2_biotin_el_hl_a. (cell line) | GSM359175(GSM359175) hela_5_pct. (hela) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | GSM359177(GSM359177) hela_nucl_a. (hela) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR037936(GSM510474) 293cand1. (cell line) | GSM359210(GSM359210) hepg2_biotin_el_hl_b. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR040011(GSM532896) G529T. (cervix) | SRR029129(GSM416758) SW480. (cell line) | DRR001484(DRX001038) "Hela long nuclear cell fraction, control". (hela) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM532874(GSM532874) G699T. (cervix) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | GSM359186(GSM359186) hepg2_untrtd. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR029127(GSM416756) A549. (cell line) | GSM359185(GSM359185) hepg2_tap_pasr. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR029130(GSM416759) DLD2. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577589(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | GSM359181(GSM359181) hela_phil_smrna_nuc. (hela) | GSM359195(GSM359195) HepG2_TAP_depl. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................GTGCTTTCTGTCCCTCCCAGGTCTG............................................. | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |