| (4) B-CELL | (4) BREAST | (21) CELL-LINE | (3) CERVIX | (1) FIBROBLAST | (2) HEART | (2) HELA | (4) LIVER | (1) OTHER | (1) RRP40.ip | (4) SKIN | (1) TESTES | (1) UTERUS | (1) XRN.ip |
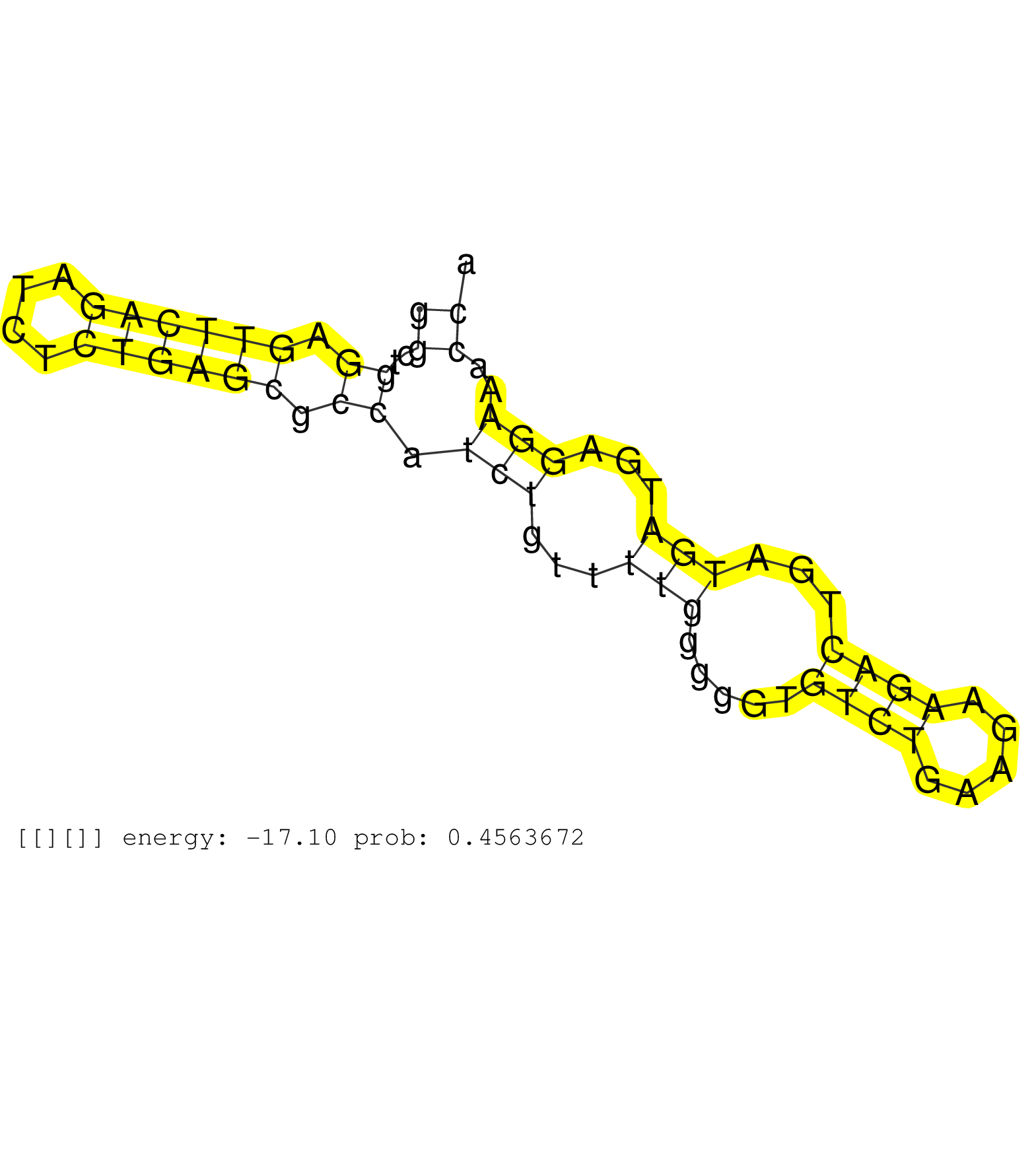
| AGCAGGAGAGACAGCGAGAAGAGCTTCTGTCTCGAACCTTCACCACTAACGTAAGCCAGGCCCGTGGTGAGGGTCGGCCTGCACTTGACAGATCGTGGGGGCTGGAGTTCAGATCTCTGAGCGCCATCTGTTTTGGGGGTGTCTGAAGAAGACTGATGATGAGGAAACCAACTGGAAATGACAGGAACTTTGTGCATTGCCACAGAGACTAGACCTAGACAGAGAATCCCTTTTTCTAAGACAAAAACCT ...................................................................................................((.(((.((((((....)))))).))).))...........((((.....))))................................................................................................. ...................................................................................................100.................................................................167................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189782 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR553576(SRX182782) source: Testis. (testes) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR037936(GSM510474) 293cand1. (cell line) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR191568(GSM715678) 51genomic small RNA (size selected RNA from t. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR029129(GSM416758) SW480. (cell line) | GSM532875(GSM532875) G547N. (cervix) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | GSM359196(GSM359196) m3m7G_ip. (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR040028(GSM532913) G026N. (cervix) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................GTGTCTGAAGAAGACTGATGATGAG....................................................................................... | 25 | 1 | 10.00 | 10.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTCTGAAGAAGACTGATGATGAGGAA.................................................................................... | 28 | 1 | 8.00 | 8.00 | 1.00 | - | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTCTGAAGAAGACTGATGATGAGG...................................................................................... | 26 | 1 | 7.00 | 7.00 | 2.00 | - | - | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTCTGAAGAAGACTGATGATGAGGA..................................................................................... | 27 | 1 | 5.00 | 5.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTCTGAAGAAGACTGATGATGAGGAAA................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................GAGTTCAGATCTCTGTGG................................................................................................................................ | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................CAGATCTCTGAGCGCCATCTGTT...................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GGTGTCTGAAGAAGACTGATGATGAG....................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TTCACCACTAACGTAAGCCAGGCCCGTGGTGAGGGT................................................................................................................................................................................ | 36 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................................CCACAGAGACTAGACCTAGACAGAG.......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................AGAAGAGCTTCTGTCTCGAACCTTCACC.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..CAGGAGAGACAGCGAGAAGAGCTTCTGTCCCGA....................................................................................................................................................................................................................... | 33 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................GTCGGCCTGCACTTGACAGATCGTGGGG...................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTCTGAAGAAGACTGATGATGAGGAAAA.................................................................................. | 30 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ACAGGAACTTTGTGCATTGCCACAG............................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GACAGCGAGAAGAGCTTCTGAA........................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................ACGTAAGCCAGGCCCGGCG....................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................AAACCAACTGGAAATGACAGGAACTTTG.......................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGTCTGAAGAAGACTGATGATGAGG...................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTGTCTGAAGAAGACTGATGATG......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................AAACCAACTGGAAATGACAGGAACT............................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................AAGACTGATGATGAGGAAACC................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CAACTGGAAATGACAGGAACTTTGTGC....................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................ACTTGACAGATCGTGGGGGCTGGGG............................................................................................................................................... | 25 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................ACTTGACAGATCGTGGGGGCTGGAG............................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GTCTGAAGAAGACTGATGATGAGG...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGACTAGACCTAGACAGAGAATCCCTTCCTC.............. | 31 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................AAGAAGACTGATGATGAGGAAACCAACTG............................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGCACTTGACAGATCG........................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................ACTGATGATGAGGAAACCAACTGGAAA........................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................GTCTCGAACCTTCACCACTAACGACT.................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................AGACTGATGATGAGGGGCT.................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................CTTCTGTCTCGAACCTTCACCACTAAC........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................AGAGACTAGACCTAGACAGAGAATCCCTTTCC............... | 32 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................CTTCTGTCTCGAACCTTCACC.............................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCTGAAGAAGACTGATGATGAG....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................AAACCAACTGGAAATGACAGGAAC.............................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GCGAGAAGAGCTTCTGTCTCGAACC.................................................................................................................................................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ..............................................................................................................................TCTGTTTTGGGGGTGTC........................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............AGCGAGAAGAGCTTCTGTC........................................................................................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GACAGCGAGAAGAGCTTCTGT............................................................................................................................................................................................................................ | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...AGGAGAGACAGCGAGAAGA.................................................................................................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...............GAGAAGAGCTTCTGTCTC......................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ................AGAAGAGCTTCTGTCTCGAACC.................................................................................................................................................................................................................... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ..............................................................................................................................................................................................TGTGCATTGCCACAGAGACTAGACCTAGA............................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................GAGTTCAGATCTCTGAG................................................................................................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| AGCAGGAGAGACAGCGAGAAGAGCTTCTGTCTCGAACCTTCACCACTAACGTAAGCCAGGCCCGTGGTGAGGGTCGGCCTGCACTTGACAGATCGTGGGGGCTGGAGTTCAGATCTCTGAGCGCCATCTGTTTTGGGGGTGTCTGAAGAAGACTGATGATGAGGAAACCAACTGGAAATGACAGGAACTTTGTGCATTGCCACAGAGACTAGACCTAGACAGAGAATCCCTTTTTCTAAGACAAAAACCT ...................................................................................................((.(((.((((((....)))))).))).))...........((((.....))))................................................................................................. ...................................................................................................100.................................................................167................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189782 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR553576(SRX182782) source: Testis. (testes) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR037936(GSM510474) 293cand1. (cell line) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR191568(GSM715678) 51genomic small RNA (size selected RNA from t. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR029129(GSM416758) SW480. (cell line) | GSM532875(GSM532875) G547N. (cervix) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | GSM359196(GSM359196) m3m7G_ip. (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR040028(GSM532913) G026N. (cervix) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................AGATCGTGGGGGCTGAAA............................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |