| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (1) AGO2.ip | (1) AGO3.ip | (4) B-CELL | (5) BREAST | (15) CELL-LINE | (1) FIBROBLAST | (7) HEART | (4) HELA | (2) LIVER | (3) OTHER | (19) SKIN | (1) TESTES | (1) XRN.ip |
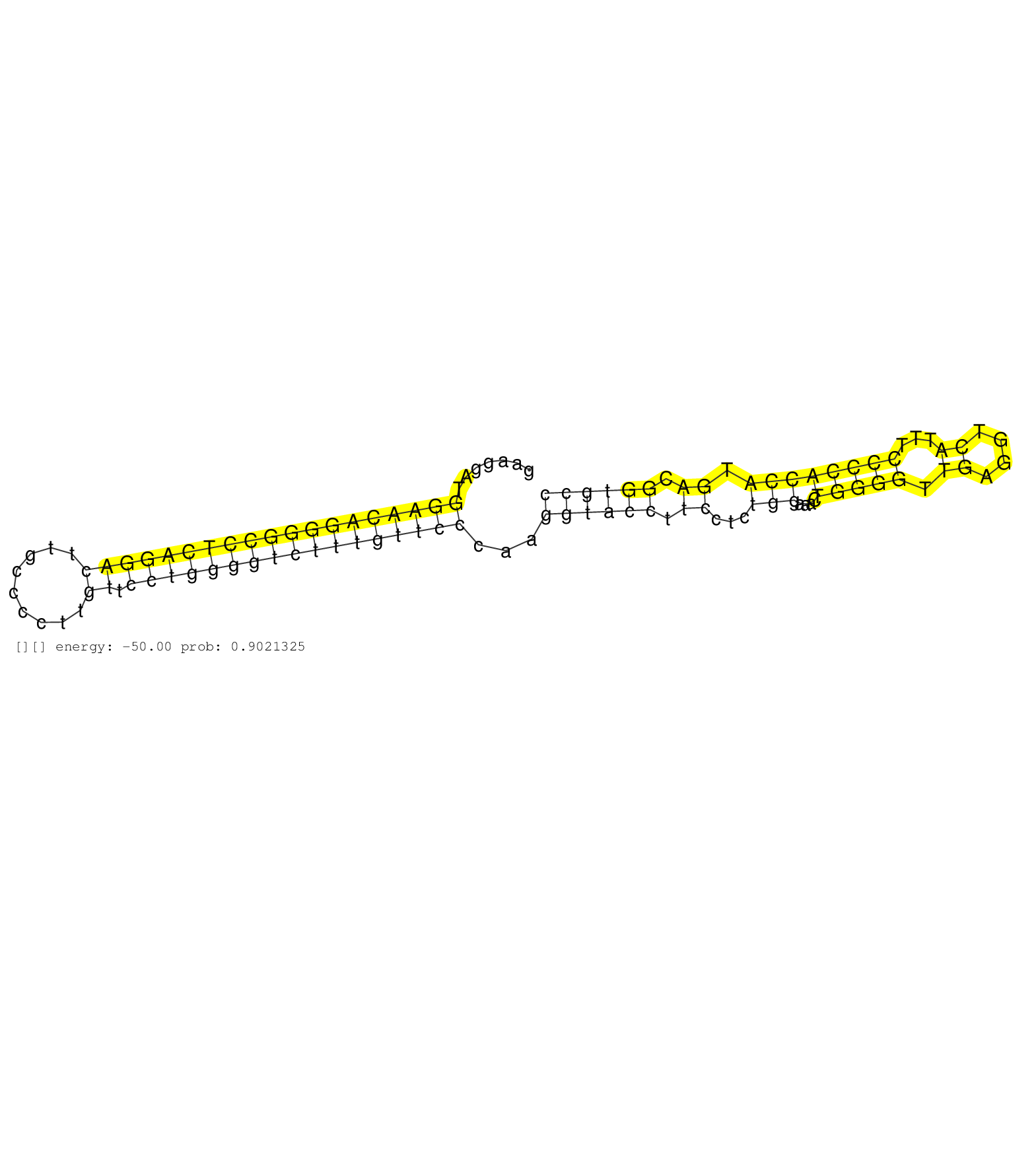
| CCTGGATGGCTGTGTTCAATGCCATGCCCACCATCTGCTTCGGATTTCAGGTGCCAGTGCCAGTTGCAAGGTCCCTGCTGTCCCCTTAGATCACCCCACCCTGGGATGGAACCCAGCAAGGAGGAGGAACCCAGGCAGAATGGAGGAAATCCTGGGGGGAAGGATGGAACAGGGGCCTCAGGACTTGCCCCTTGTTCCTGGGGTCTTTGTTCCCAAGGTACCTTCCTCTGGAAACTGGGGTTGAGGTCATTTCCCCACCATGACGGTGCCCTTCACCTGGACAGTGCCACGTCAGCAGTGTGCCCGTCTTCAACAGCATGCAGCAGCCTGAAGT ....................................................................................................(((((......)))))((((...((((..(((...((...))......(((((.......)))))......))).))))....))))................................................................................................................................................... ................................................................................................97........................................................................................187................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR553574(SRX182780) source: Heart. (Heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM416733(GSM416733) HEK293. (cell line) | SRR189782 | SRR038855(GSM458538) D10. (cell line) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR037931(GSM510469) 293GFP. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | TAX577590(Rovira) total RNA. (breast) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR029126(GSM416755) 143B. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR553576(SRX182782) source: Testis. (testes) | TAX577579(Rovira) total RNA. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038853(GSM458536) MELB. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGA....................................................................................................................................................... | 20 | 1 | 12.00 | 12.00 | 9.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGACT..................................................................................................................................................... | 22 | 1 | 12.00 | 12.00 | 5.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................GATGGAACAGGGGCCTCAGG........................................................................................................................................................ | 20 | 1 | 8.00 | 8.00 | - | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................ATCTGCTTCGGATTTCAG............................................................................................................................................................................................................................................................................................ | 18 | 1 | 8.00 | 8.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGAC...................................................................................................................................................... | 21 | 1 | 5.00 | 5.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................CTGCTTCGGATTTCAG............................................................................................................................................................................................................................................................................................ | 16 | 1 | 5.00 | 5.00 | - | - | - | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................TCTGCTTCGGATTTCAG............................................................................................................................................................................................................................................................................................ | 17 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GATGGAACAGGGGCCTCAGGAA...................................................................................................................................................... | 22 | 1 | 3.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGGAACAGGGGCCTCAGGATT..................................................................................................................................................... | 21 | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................ATTTCAGGTGCCAGTGCCAGT.............................................................................................................................................................................................................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGGAACAGGGGCCTCAGGAT...................................................................................................................................................... | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................................CTGGGGTTGAGGTCATTTCCCCACCATGACGG.................................................................... | 32 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGAG...................................................................................................................................................... | 21 | 1 | 2.00 | 12.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................CCACGTCAGCAGTGTGCC.............................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGACTAAT.................................................................................................................................................. | 25 | 1 | 1.00 | 12.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGGGATGGAACCCAGTCTC...................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGG........................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TCTGCTTCGGATTTCAGGTG......................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGATAT.................................................................................................................................................... | 23 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGAT...................................................................................................................................................... | 21 | 1 | 1.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................AGGAGGAGGAACCCAGGCAGA................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CCATCTGCTTCGGATTTCAGG........................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GATGGAACAGGGGCCTCAGGA....................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGGAACAGGGGCCTCAGGAAGA.................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGATT..................................................................................................................................................... | 22 | 1 | 1.00 | 12.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................ATGCCATGCCCACCATCTGCTTC..................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................CCCCTTGTTCCTGGGGTCTTTGTTC.......................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TGGAAACTGGGGTTGAGGTTTCC................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................CATGCCCACCATCTGCTTCGGATTTCAG............................................................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................GAGGAACCCAGGCAGAATGGAGGAAATCCTG.................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................GGATGGAACAGGGGCCTCAGGA....................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGGAACAGGGGCCTCAGGACTTGC.................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................AGGAGGAACCCAGGCAGAATGGAGG............................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGACTT.................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGAACAGGGGCCTCAGGAAA..................................................................................................................................................... | 22 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CCCAGCAAGGAGGAGGAACCCAGGC...................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGGAACAGGGGCCTCAGGACT..................................................................................................................................................... | 21 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| CCTGGATGGCTGTGTTCAATGCCATGCCCACCATCTGCTTCGGATTTCAGGTGCCAGTGCCAGTTGCAAGGTCCCTGCTGTCCCCTTAGATCACCCCACCCTGGGATGGAACCCAGCAAGGAGGAGGAACCCAGGCAGAATGGAGGAAATCCTGGGGGGAAGGATGGAACAGGGGCCTCAGGACTTGCCCCTTGTTCCTGGGGTCTTTGTTCCCAAGGTACCTTCCTCTGGAAACTGGGGTTGAGGTCATTTCCCCACCATGACGGTGCCCTTCACCTGGACAGTGCCACGTCAGCAGTGTGCCCGTCTTCAACAGCATGCAGCAGCCTGAAGT ....................................................................................................(((((......)))))((((...((((..(((...((...))......(((((.......)))))......))).))))....))))................................................................................................................................................... ................................................................................................97........................................................................................187................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR553574(SRX182780) source: Heart. (Heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM416733(GSM416733) HEK293. (cell line) | SRR189782 | SRR038855(GSM458538) D10. (cell line) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR037931(GSM510469) 293GFP. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | TAX577590(Rovira) total RNA. (breast) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR029126(GSM416755) 143B. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR553576(SRX182782) source: Testis. (testes) | TAX577579(Rovira) total RNA. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038853(GSM458536) MELB. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................TTAGATCACCCCACCAA........................................................................................................................................................................................................................................ | 17 | 8.00 | 0.00 | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .....................................................................................TTAGATCACCCCACCCAA....................................................................................................................................................................................................................................... | 18 | 6.00 | 0.00 | - | - | - | - | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................TTAGATCACCCCACCCAC....................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................GGTGCCAGTGCCAGTAT............................................................................................................................................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................TGACGGTGCCCTTCACTGT....................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................CTGGGATGGAACCCAGCGG....................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................GGAAGGATGGAACAGAGC............................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................GATGGAACCCAGCAAGAG.................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| .........................................................................................TCCCAGGGTGGGGTGAT.................................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |