| (1) AGO2.ip | (1) AGO3.ip | (5) B-CELL | (12) BRAIN | (7) BREAST | (21) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (8) HEART | (3) HELA | (1) KIDNEY | (6) LIVER | (1) OTHER | (28) SKIN | (1) TESTES | (2) UTERUS |
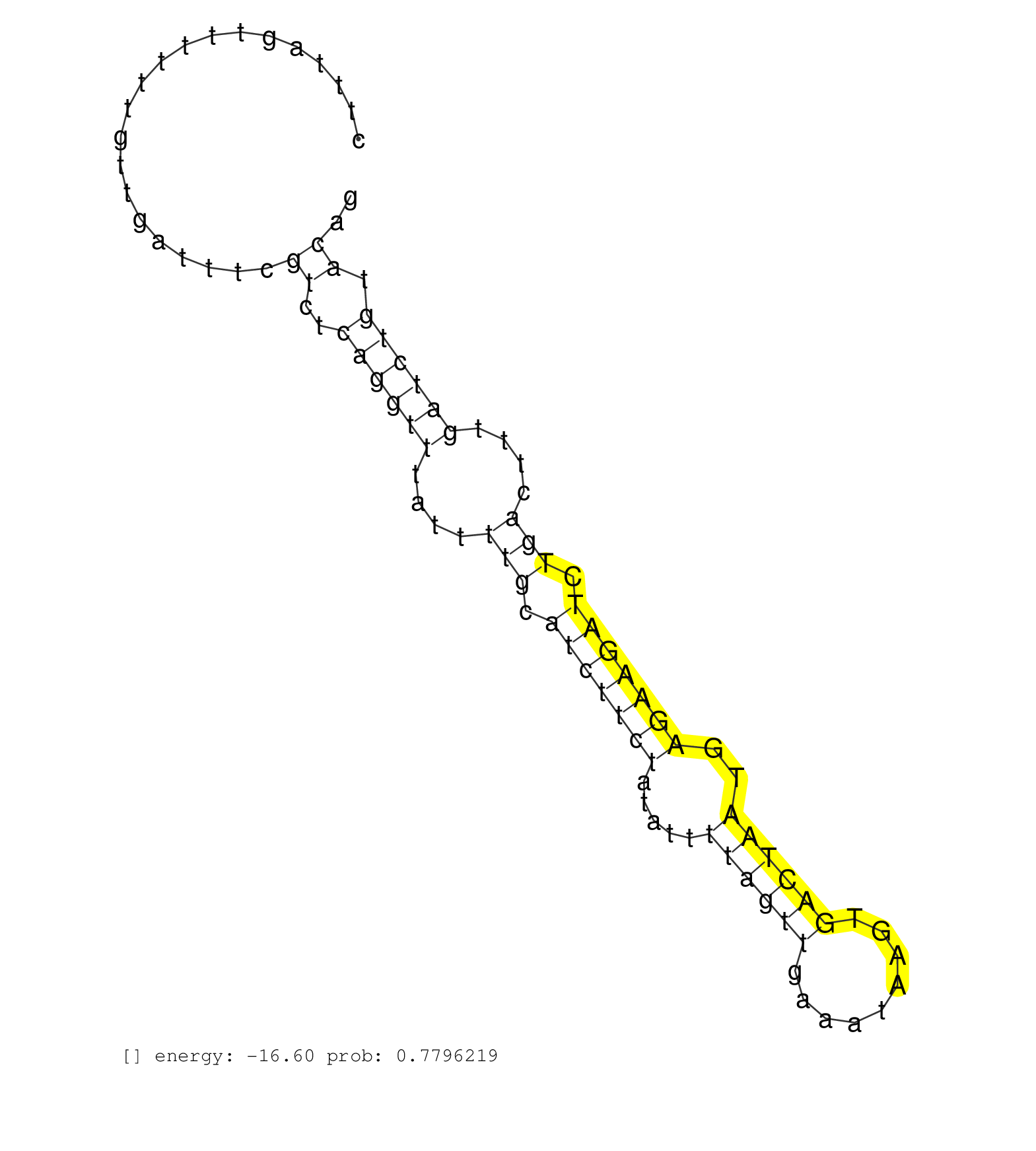
| AAGATTGCCTTTTCCAAAGATCTTGATGATTGTTTGACAGTTCTGCTTAGAGCCAAGTGAAATTTTTCCATTTGGGCTTCCGAGTAACAGTGTGGGTTAGCTTTAGTTTTTTGTTGATTTCGTCTCAGGTTTATTTTGCATCTTCTATATTTTAGTTGAAATAAGTGACTAATGAGAAGATCTGACTTTGATCTGTACAGGCCTCTCTATGTCACTGTAGTGTATCAAGGCAAATGTTCTCACTGCATGA ................................................................................................................(((((((...(((((((((...((((..((..(((.....)))..))..)))).))))..))))).)))).)))................................................................ ....................................................................................................101.....................................................................................189........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553576(SRX182782) source: Testis. (testes) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR553575(SRX182781) source: Kidney. (Kidney) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR343337 | SRR553574(SRX182780) source: Heart. (Heart) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR191410(GSM715520) 20genomic small RNA (size selected RNA from t. (breast) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR191403(GSM715513) 44genomic small RNA (size selected RNA from t. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR029131(GSM416760) MCF7. (cell line) | SRR191538(GSM715648) 49genomic small RNA (size selected RNA from t. (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................AAGTGACTAATGAGAAGATCTGA................................................................. | 23 | 1 | 20.00 | 20.00 | - | - | 1.00 | - | 1.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATCTG.................................................................. | 23 | 1 | 16.00 | 16.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATCT................................................................... | 22 | 1 | 16.00 | 16.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - |
| ..................................................................................................................................................................AAGTGACTAATGAGAAGATCT................................................................... | 21 | 1 | 14.00 | 14.00 | - | - | - | - | - | - | - | - | 2.00 | 2.00 | - | - | - | 2.00 | - | 1.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................GGCCTCTCTATGTCACTGTAG.............................. | 21 | 1 | 11.00 | 11.00 | - | - | 2.00 | - | 1.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AAGTGACTAATGAGAAGATCTG.................................................................. | 22 | 1 | 11.00 | 11.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATC.................................................................... | 21 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATCTGA................................................................. | 24 | 1 | 7.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATCTGT................................................................. | 24 | 1 | 7.00 | 16.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................ATAAGTGACTAATGAGAAGATCT................................................................... | 23 | 1 | 6.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGAT..................................................................... | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................ATAAGTGACTAATGAGAAGATCTG.................................................................. | 24 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AAGTGACTAATGAGAAGATCTGT................................................................. | 23 | 1 | 4.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................ATAAGTGACTAATGAGAAGATC.................................................................... | 22 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGA...................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCTATGTCACTGTAGTGTATCAAGGC................... | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................ATAAGTGACTAATGAGAAGATA.................................................................... | 22 | 1 | 3.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCACTGTAGTGTATCAAGGCAAATG.............. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TCTCTATGTCACTGTAGTGTATCAAGGC................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGTGACTAATGAGAAGAT..................................................................... | 18 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGTGTGGGTTAGCTTTAGTTTTTTGTTGATC................................................................................................................................... | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GGCCTCTCTATGTCACTGTAGT............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATCTA.................................................................. | 23 | 1 | 1.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AGATCTGACTTTGATCTGTACAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ATGTCACTGTAGTGTATCAAGGC................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AAGTGACTAATGAGAAGATGG................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGTAGTGTATCAAGGC................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CCTCTCTATGTCACTGTA............................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ATGTCACTGTAGTGTATC........................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................ATAAGTGACTAATGAGAAGAT..................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGTCACTGTAGTGTATCAAGGCAAATGT............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GTGACTAATGAGAAGATCTGACTT.............................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATTTG.................................................................. | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AAGTGACTAATGAGAAGAT..................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CACTGTAGTGTATCAAGGCAAATG.............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CTATGTCACTGTAGTGTATCAAGGC................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGTGACTAATGAGAAGATC.................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGTGACTAATGAGAAGATCTGA................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGTGACTAATGAGAAGATCTGACTTTGAT.......................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AATAAGTGACTAATGAGAAGAT..................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGACTAATGAGAAGATCT................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ATGTCACTGTAGTGTATCAAGGCAAAT............... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGTGACTAATGAGAAGATCTGACTTT............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ATGTCACTGTAGTGTATCAAGGCAAATGTTC........... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AAGTGACTAATGAGAAGATATGA................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GTTTTTTGTTGATTTCGTCTCAGG......................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGTGACTAATGAGAAGATCTT.................................................................. | 23 | 1 | 1.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............CCAAAGATCTTGATGA............................................................................................................................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| AAGATTGCCTTTTCCAAAGATCTTGATGATTGTTTGACAGTTCTGCTTAGAGCCAAGTGAAATTTTTCCATTTGGGCTTCCGAGTAACAGTGTGGGTTAGCTTTAGTTTTTTGTTGATTTCGTCTCAGGTTTATTTTGCATCTTCTATATTTTAGTTGAAATAAGTGACTAATGAGAAGATCTGACTTTGATCTGTACAGGCCTCTCTATGTCACTGTAGTGTATCAAGGCAAATGTTCTCACTGCATGA ................................................................................................................(((((((...(((((((((...((((..((..(((.....)))..))..)))).))))..))))).)))).)))................................................................ ....................................................................................................101.....................................................................................189........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553576(SRX182782) source: Testis. (testes) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR553575(SRX182781) source: Kidney. (Kidney) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR343337 | SRR553574(SRX182780) source: Heart. (Heart) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR191410(GSM715520) 20genomic small RNA (size selected RNA from t. (breast) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR191403(GSM715513) 44genomic small RNA (size selected RNA from t. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR029131(GSM416760) MCF7. (cell line) | SRR191538(GSM715648) 49genomic small RNA (size selected RNA from t. (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................TAACAGTGTGGGTTAAGA.................................................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................GAGCCAAGTGAAATTGGT....................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ....................................................CCAAGTGAAATTTTTGAA.................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................GACAGTTCTGCTTAGGCTG.................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................AGTTGAAATAAGTGATAA............................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ........................................................................................................................................................................................................ACAGTGACATAGAGAGGC................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |