| (1) AGO2.ip | (2) B-CELL | (2) BRAIN | (2) BREAST | (8) CELL-LINE | (4) CERVIX | (5) HEART | (4) LIVER | (2) OTHER | (6) SKIN |
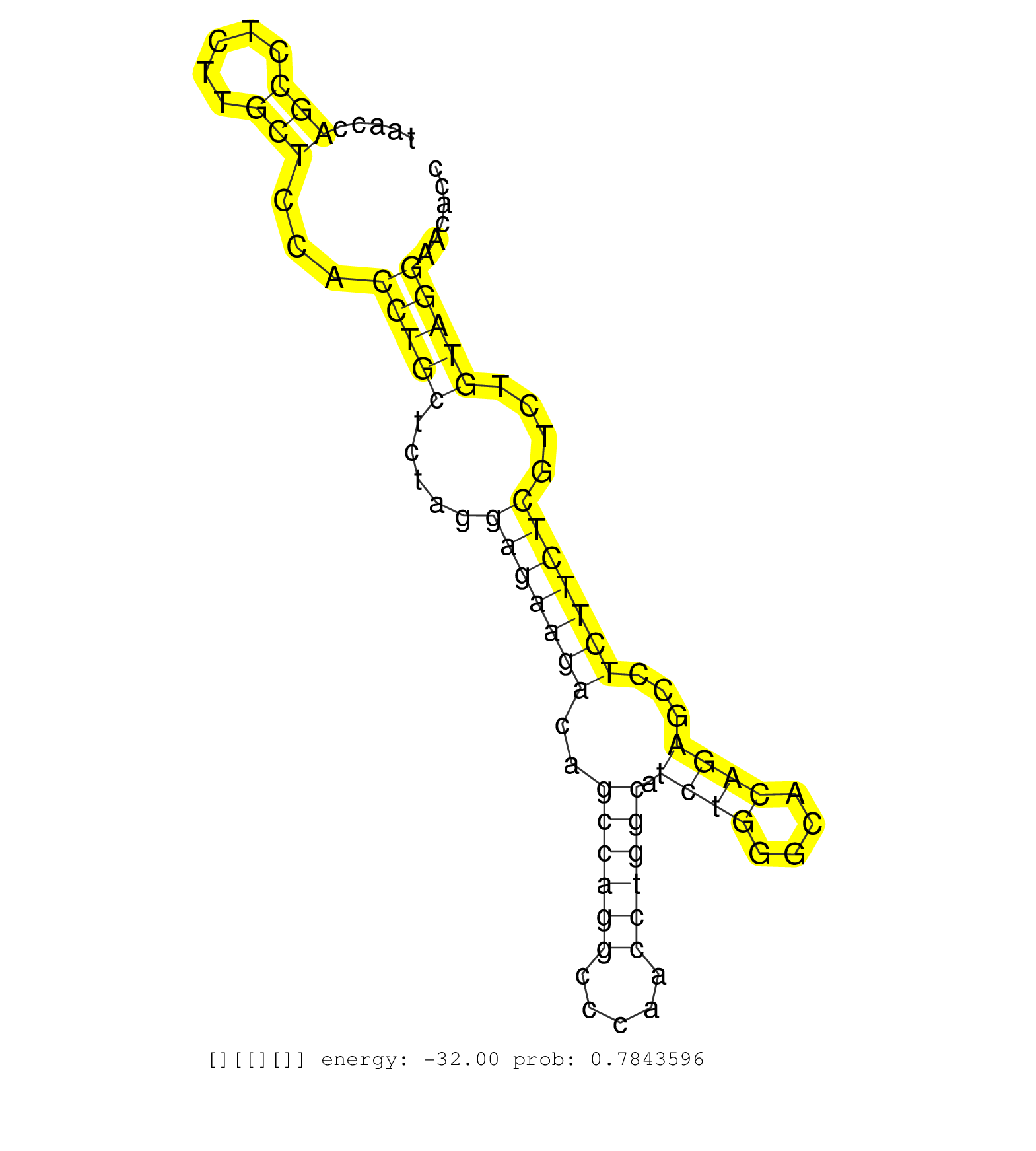
| GGGGAAGTGGCAGCACCTCCTGAAGCAGTTGGACTTTCATCACCCTACCTCTGCATCTGCCTGAAGGACAGATTTAGCCAATTAACCTAAGGTTACCTTCCTCTCTGATAAATTCCCCATTCTGTCTTCCCATGTGTTGTGTCTCGTTTTTTTCCTCCTCCTTCCCTCTTCCTTGCCCCCTCTTCCCCTAAACCTTACAGATAGCTCGTGAGGCTGAGGCAGCCATGTTCCACCGCAAGCTGTTTGAAGA ..............................................................(((((((((((.((((((..(((((...)))))...........((((............))))......)).)))).)))).))))))).................................................................................................. .......................................................56...............................................................................................................................185............................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189784 | SRR037935(GSM510473) 293cand3. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040009(GSM532894) G727T. (cervix) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR040007(GSM532892) G601T. (cervix) | SRR040039(GSM532924) G531T. (cervix) | SRR390723(GSM850202) total small RNA. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR040008(GSM532893) G727N. (cervix) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCCGCC..................................................................... | 21 | 29.00 | 0.00 | 20.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCCGC...................................................................... | 20 | 7.00 | 0.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCCGCCC.................................................................... | 22 | 5.00 | 0.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................TTCCCTCTTCCTTGCCCGCCC.................................................................... | 21 | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCTCGC...................................................................... | 20 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................CTGAAGGACAGATTTAGCC........................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....AGTGGCAGCACCTCCTGAAGCAGTTGGACTC...................................................................................................................................................................................................................... | 31 | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCCGCT..................................................................... | 21 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................CCGCAAGCTGTTTGAAGA | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCTGCC..................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GATAGCTCGTGAGGCTGAGGC.............................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TCATCACCCTACCTCCGG.................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................TGAAGGACAGATTTAGC............................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ATAGCTCGTGAGGCTGAGG............................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATAGCTCGTGAGGCTGAGGCAGCC.......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................ATGTTCCACCGCAAGC.......... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ATAGCTCGTGAGGCTGAGGCAGCCATG....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTCCCTCTTCCTTGCCCGCC..................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCAGCC..................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................CTCGTGAGGCTGAGGCA............................. | 17 | 0 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTTTTTTCCTCCTCCTTCCCCC.................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGTCC........................................................................ | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCTGCT..................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTTCCCCTAAACCTTACAGATAGCT............................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CCACCGCAAGCTGTT...... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTGAAGCAGTTGGACTTT..................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..GGAAGTGGCAGCACCTCCTGAAGCAGTTGGA......................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTCCCTCTTCCTTGCCCTCCA.................................................................... | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GATAGCTCGTGAGGCTGAGGCAGC........................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCCGCGT.................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................GCAGCCATGTTCCACCGCAAGCT......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CCTAAGGTTACCTTCCGAAC................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................CCCCTAAACCTTACAGATAGCTC........................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GTGAGGCTGAGGCAGCCATGTTCCACC................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGGCAGCACCTCCTGAAGCAGTTGGACTCTC.................................................................................................................................................................................................................... | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CTTCCCTCTTCCTTGCCATCC..................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................GTCTCGTTTTTTTCCC.............................................................................................. | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................GCTCGTGAGGCTGAGGCAGGC.......................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................TTCCCTCTTCCTTGCCCGC...................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......TGGCAGCACCTCCTGGGAC................................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GATAGCTCGTGAGGCTGAGGCAGCCATGT...................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCGTGAGGCTGAGGCGGA........................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................TTCCCTCTTCCTTGCCCTCCC.................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................AGCTCGTGAGGCTGAGGC.............................. | 18 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........AGCACCTCCTGAAGCA............................................................................................................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................AAATTCCCCATTCTGTC............................................................................................................................ | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| ............................................................................................................................TCTTCCCATGTGTTGT.............................................................................................................. | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - |
| GGGGAAGTGGCAGCACCTCCTGAAGCAGTTGGACTTTCATCACCCTACCTCTGCATCTGCCTGAAGGACAGATTTAGCCAATTAACCTAAGGTTACCTTCCTCTCTGATAAATTCCCCATTCTGTCTTCCCATGTGTTGTGTCTCGTTTTTTTCCTCCTCCTTCCCTCTTCCTTGCCCCCTCTTCCCCTAAACCTTACAGATAGCTCGTGAGGCTGAGGCAGCCATGTTCCACCGCAAGCTGTTTGAAGA ..............................................................(((((((((((.((((((..(((((...)))))...........((((............))))......)).)))).)))).))))))).................................................................................................. .......................................................56...............................................................................................................................185............................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189784 | SRR037935(GSM510473) 293cand3. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040009(GSM532894) G727T. (cervix) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR040007(GSM532892) G601T. (cervix) | SRR040039(GSM532924) G531T. (cervix) | SRR390723(GSM850202) total small RNA. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR040008(GSM532893) G727N. (cervix) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................TTTTTTCCTCCTCCTTCTT.................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ..................................TTTCATCACCCTACCAAGA..................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................AATTCCCCATTCTGTAAGG......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................TTTTTTTCCTCCTCCTACA..................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................CTTTCATCACCCTACAGA....................................................................................................................................................................................................... | 18 | 0.22 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | 0.11 | - | - | |
| .................................GTAGGGTGATGAAAG.......................................................................................................................................................................................................... | 15 | 9 | 0.22 | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.22 | - | - | - | - | - | - |
| .................................CTTTCATCACCCTACAATC...................................................................................................................................................................................................... | 19 | 0.11 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | |
| .................................CTTTCATCACCCTACAGAA...................................................................................................................................................................................................... | 19 | 0.11 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | |
| .................................CTTTCATCACCCTACAG........................................................................................................................................................................................................ | 17 | 0.11 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - |