| (1) B-CELL | (5) BRAIN | (3) BREAST | (23) CELL-LINE | (1) HEART | (4) HELA | (3) LIVER | (2) OTHER | (3) SKIN | (1) TESTES |
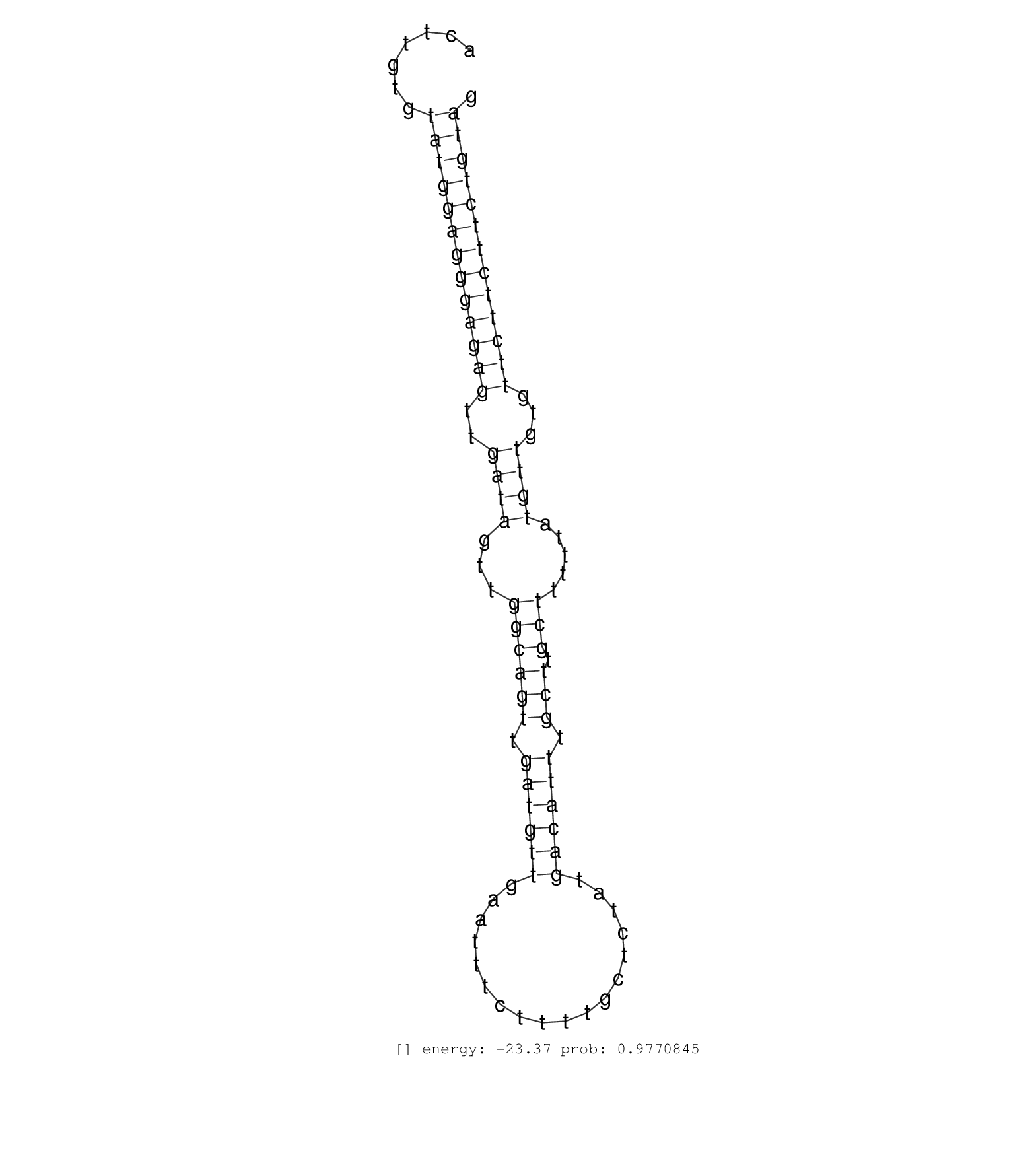
| CTCCCCTATAATGAGCATTGTGTTGCTAACAGGAAATCTGAAGTCATGATACCATGATGTAAATTCTCAAACATCTGAGTAAAAGTGATCATGACCAGAAACTTGTGTATGGAGGGAGAGTTGATAGTTGGCAGTTGATGTTGAATTTCTTTTGCTCTATGACATTTGCTTGCTTTTTATGTTGTGTTCTTCTTCTGTAGACTGTGAAGGGGATGTTGGACAATCCTAATGAACCTGTTAGTGACCTCTC ...................................................(((...(((.(((((((.....((((..((.((((..((.......)).)))).))..))))..))))))).)))..)))....................................................................................................................... ............................................45.................................................................................................144........................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR037943(GSM510481) 293DcrTN. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM416733(GSM416733) HEK293. (cell line) | SRR189782 | SRR029128(GSM416757) H520. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553576(SRX182782) source: Testis. (testes) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM359174(GSM359174) hela_5_1. (hela) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM359176(GSM359176) hela_chip_smrna_pure. (hela) | SRR191442(GSM715552) 107genomic small RNA (size selected RNA from . (breast) | SRR029130(GSM416759) DLD2. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577453(Rovira) total RNA. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................TACCATGATGTAAATTCTCAAACATCTGAGT.......................................................................................................................................................................... | 31 | 1 | 118.00 | 118.00 | 14.00 | 23.00 | 12.00 | 15.00 | 13.00 | 12.00 | - | 3.00 | 3.00 | 3.00 | - | 3.00 | 3.00 | 3.00 | - | 3.00 | - | 1.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ................................................ATACCATGATGTAAATTCTCAAACATCTGAGT.......................................................................................................................................................................... | 32 | 1 | 95.00 | 95.00 | 35.00 | 28.00 | 9.00 | - | 1.00 | - | 5.00 | 4.00 | 2.00 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ................................................ATACCATGATGTAAATTCTCAAACATCT.............................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................ACCATGATGTAAATTCTCAAACATCTGAGT.......................................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAAGGGGATGTTGGACA............................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TACCATGATGTAAATGTTG...................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................ATACCATGATGTAAATTCTCAAACAT................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGACAATCCTAATGAACCTGTTAGT........ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGACAATCCTAATGAACCTGTTAGT........ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TACCATGATGTAAATTCTCAAACATA............................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TACCATGATGTAAATTCTCAAACATCTGAGG.......................................................................................................................................................................... | 31 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................ATACCATGATGTAAATTCTCAAACA................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................ATGAACCTGTTAGTGACC.... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TACCATGATGTAAATTCTCAAACAT................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TACCATGATGTAAATGTAG...................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................GGGGATGTTGGACAATC......................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGATAGTTGGCAGTTAATC.............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................ATACCATGATGTAAATTCTCAAACATCTGATA.......................................................................................................................................................................... | 32 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................TGTAGACTGTGAAGGCATG.................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .............................................................................................................................................................................................................GAAGGGGATGTTGGACAA........................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ACTGTGAAGGGGATGTTGGAC............................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGACTGTGAAGGGGATGTT................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................GTAAATTCTCAAACATCTGAGT.......................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGATAGTTGGCAGTTAATA.............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................ATACCATGATGTAAATTCTCAAACATC............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TAATGAACCTGTTAGTGAC..... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................ATACCATGATGTAAATTCTCA..................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGACTGTGAAGGGGATGTTGGACAATCCTAATG................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GTTGGACAATCCTAATCA.................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................ACTGTGAAGGGGATGTTGGA.............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................GTGAAGGGGATGTTGGACA............................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................TGGACAATCCTAATGAACCTGTTAGTGA...... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CTGTGAAGGGGATGTTGG............................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| CTCCCCTATAATGAGCATTGTGTTGCTAACAGGAAATCTGAAGTCATGATACCATGATGTAAATTCTCAAACATCTGAGTAAAAGTGATCATGACCAGAAACTTGTGTATGGAGGGAGAGTTGATAGTTGGCAGTTGATGTTGAATTTCTTTTGCTCTATGACATTTGCTTGCTTTTTATGTTGTGTTCTTCTTCTGTAGACTGTGAAGGGGATGTTGGACAATCCTAATGAACCTGTTAGTGACCTCTC ...................................................(((...(((.(((((((.....((((..((.((((..((.......)).)))).))..))))..))))))).)))..)))....................................................................................................................... ............................................45.................................................................................................144........................................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR037943(GSM510481) 293DcrTN. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM416733(GSM416733) HEK293. (cell line) | SRR189782 | SRR029128(GSM416757) H520. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553576(SRX182782) source: Testis. (testes) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM359174(GSM359174) hela_5_1. (hela) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM359176(GSM359176) hela_chip_smrna_pure. (hela) | SRR191442(GSM715552) 107genomic small RNA (size selected RNA from . (breast) | SRR029130(GSM416759) DLD2. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577453(Rovira) total RNA. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................GTTCTTCTTCTGTAGCACA.............................................. | 19 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...................................................................................................................................................................ATTTGCTTGCTTTTTACGG.................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................GTTCTTCTTCTGTAGTACA.............................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................GTTGATGTTGAATTTACT................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ...........................................................................................................................................................................GCTTTTTATGTTGTGTCT............................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........AATGAGCATTGTGTTGTCT.............................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |