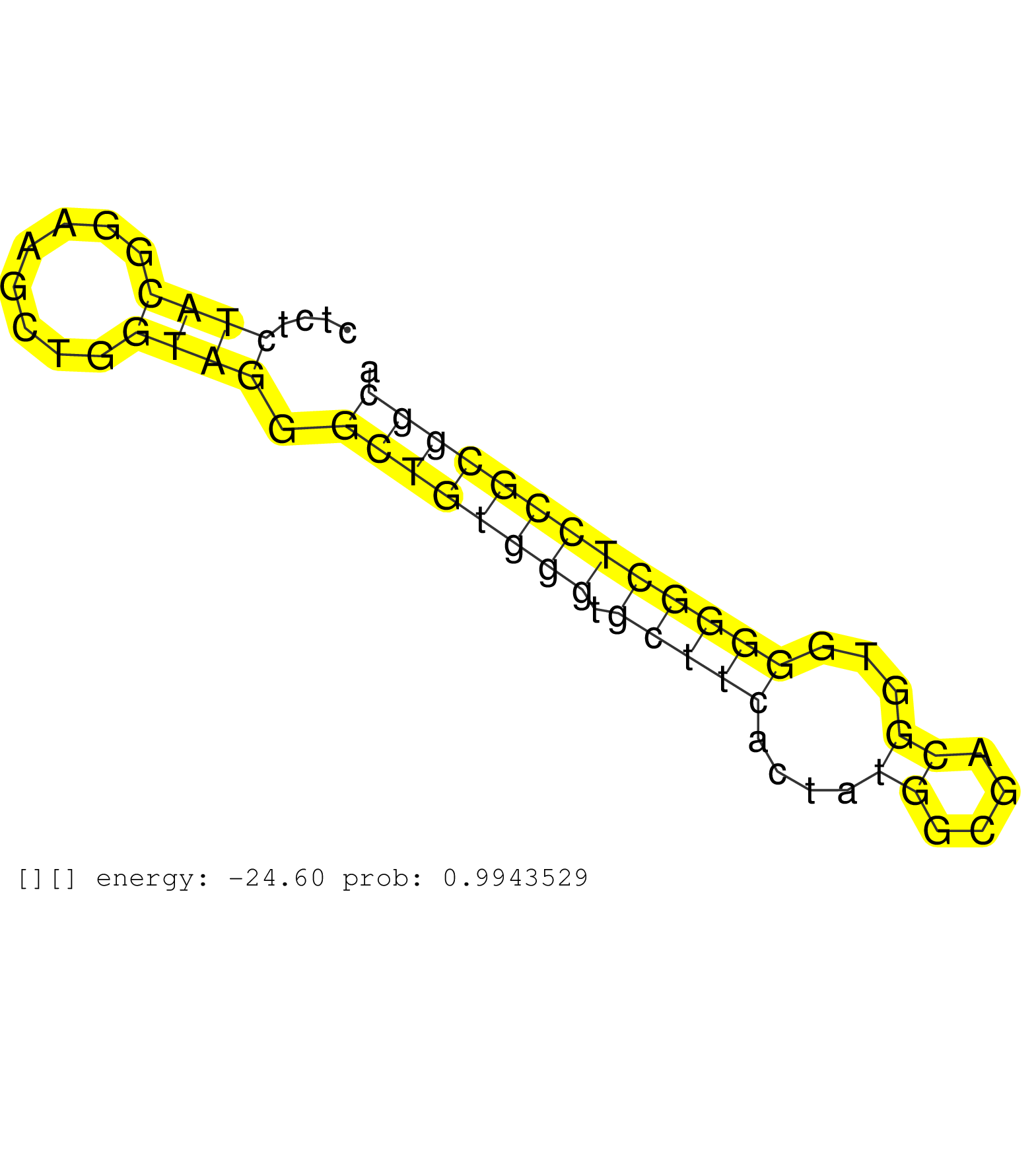
| (1) AGO1.ip | (2) AGO2.ip | (4) B-CELL | (2) BREAST | (12) CELL-LINE | (1) HEART | (2) HELA | (2) LIVER | (1) OTHER | (6) SKIN | (1) XRN.ip |
| TCGCGAGTTCCTACCACCCGGCACGGCCTGAGGTCTACGACTCACTGCAGGTACCAGCCAGCCAGAGTGCTTGAAGGCCGGTACTGGAAAGAGGAGGTAGCATCCAGGGCTGGGGGTGCTGCCTGCCCCACAGCTGGTGTGTTTTGCTCCCAGGATGCAGCTCTGGCCCCCGAGTTCCTGGCCGTGACTGAGTACAGCGTGTC ............................................................................((((((..(((.....(((((((((((((........))))))))))).)))))..))))))................................................................. ........................................................................73..............................................................................153................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189786 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR189787 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR189784 | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................AAGGCCGGTACTGGAAAGAGGAGGTAGCACC................................................................................................... | 31 | 10.00 | 0.00 | 8.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................AAGGCCGGTACTGGAAAGAGGAGGTAGCATC................................................................................................... | 31 | 1 | 6.00 | 6.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................AAGGCCGGTACTGGAAAGAGGAGGTAGCATCC.................................................................................................. | 32 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAG....................................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................AAGGCCGGTACTGGAAAGAGGAGGTAGCATT................................................................................................... | 31 | 3.00 | 0.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAGCATA................................................................................................... | 27 | 1 | 2.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TGAAGGCCGGTACTGGAAAGA............................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................CGGTACTGGAAAGAGGAGGTAG....................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................CCGGTACTGGAAAGAGGAGGGAG....................................................................................................... | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .......................................................................................................................................................AGGATGCAGCTCTGGCA................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AGGTAGCATCCAGGGCTGGGGGTG..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAGCAT.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................CGGCACGGCCTGAGGTCTA...................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CCGGTACTGGAAAGAGGAG........................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAGA...................................................................................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GTCTACGACTCACTGCAG......................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCCAGCCAGAGTGCTTGTGAT.............................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................AGGATGCAGCTCTGGCCCCCGAG............................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................AAGGCCGGTACTGGAAAGAGGAGGTAGCAC.................................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................AAGGCCGGTACTGGAAAGAGGAGGTAGCATCT.................................................................................................. | 32 | 1 | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TTGAAGGCCGGTACTGGAAAGAGGAGGTAGCAT.................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................ATGCAGCTCTGGCCCCCGAGTTCC......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CTGGAAAGAGGAGGTAGCA..................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTACCAGCCAGCCAGAGTGCTTGAAGGCCGGTA........................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TGAAGGCCGGTACTGGAAAGAGGAGGTAGCAT.................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GCCGGTACTGGAAAGAGGAGGTAGCATC................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAT....................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .........................................................................AAGGCCGGTACTGGAAAGAGGAGGTACCT..................................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAGT...................................................................................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAGC...................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............ACCACCCGGCACGGCCTGAGGTCCACG.................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................TGGTGTGTTTTGCTCCCA................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CCGGTACTGGAAAGAGGAGGTAGCATC................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TTGAAGGCCGGTACTGGAAAGAGGAGGTAGCA..................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........CCTACCACCCGGCACGGCC............................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCGCGAGTTCCTACCACCCGGCACGGCCTGAGGTCTACGACTCACTGCAGGTACCAGCCAGCCAGAGTGCTTGAAGGCCGGTACTGGAAAGAGGAGGTAGCATCCAGGGCTGGGGGTGCTGCCTGCCCCACAGCTGGTGTGTTTTGCTCCCAGGATGCAGCTCTGGCCCCCGAGTTCCTGGCCGTGACTGAGTACAGCGTGTC ............................................................................((((((..(((.....(((((((((((((........))))))))))).)))))..))))))................................................................. ........................................................................73..............................................................................153................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189786 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR189787 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR189784 | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................ACCAGCTGTGGGGCAGGCAG................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGGCCCCCGAGTTCCTGGCCGTGACAA............. | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ........GTAGACCTCAGGCCGTGCCGGGTGGTAGGA..................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....AGTTCCTACCACCCGGCGTG.................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| .......................TGAGTCGTAGACCTCAGGCCG............................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................CTCAGGCCGTGCCGGG........................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TACTCAGTCACGGCCAGGAACT......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................CCCGGCACGGCCTGAGGTCTACGACTCGG.............................................................................................................................................................. | 29 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................AGCATCCAGGGCTGGGGGCCC.................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |