| (3) B-CELL | (1) BREAST | (5) CELL-LINE | (4) CERVIX | (1) HEART | (1) OTHER | (3) SKIN |
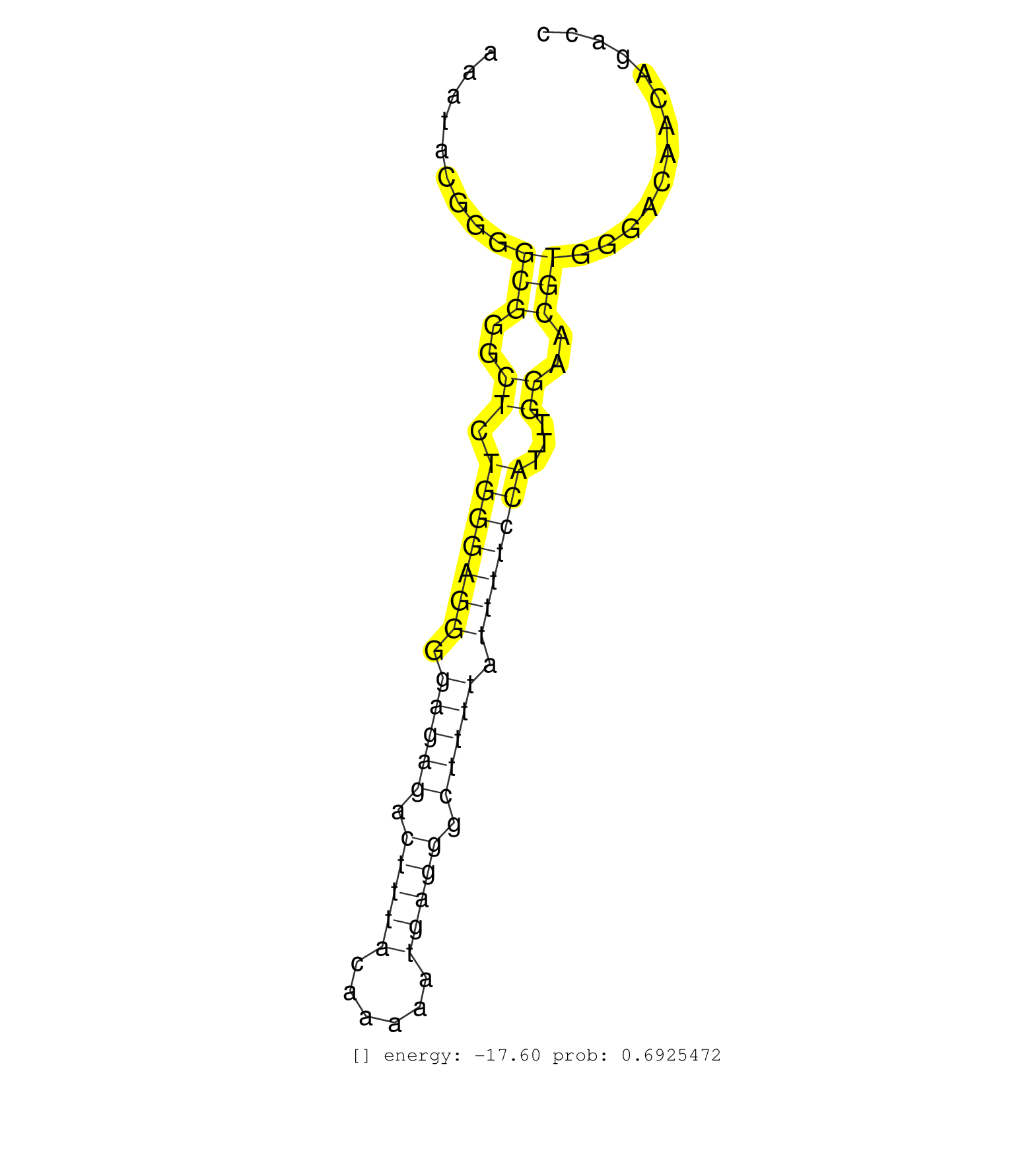
| TTCACCTGAATTACCTCGTTTAACCTGACGGTCTCTGCAAAGCCCCCATCTCCAAATAAAGATCACACTCTGAGGAATGGGGTTTAGAGCCCCCATCGTGTCTTTCTGGGGGTACACAGTTTGCTCCCTAACACCCTTTCCGGGGGTCTGGCCGTGCCTGCAGTCTCCCCATCTGACCAGTCGTGTCTGTTCCCCTGCAGAACCGGGCTGTGGGTCGGCCCAAGGGGAAGCTGGGCCCGGCCTCCGCGGT ................................................................((((.(((.((((.(((((((((.(((((((...........)))))))....((....))..)))).))))))))).((((...(((.((....)).))).)))).......)))..))))((((......)))).................................................. ........................................................57.............................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR343334 | SRR343335 | SRR343336 | SRR343337 | SRR189784 | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR040007(GSM532892) G601T. (cervix) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR038857(GSM458540) D20. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | TAX577579(Rovira) total RNA. (breast) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM532876(GSM532876) G547T. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................ATCACACTCTGAGGACG............................................................................................................................................................................ | 17 | 102.00 | 0.00 | 49.00 | 31.00 | 13.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................ATCACACTCTGAGGACGG........................................................................................................................................................................... | 18 | 19.00 | 0.00 | 7.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................ATCACACTCTGAGGACGA........................................................................................................................................................................... | 18 | 3.00 | 0.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................ATCACACTCTGAGGACGGC.......................................................................................................................................................................... | 19 | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................TCCCCTGCAGAACCGTTAA......................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................CGGGCTGTGGGTCGGCGGA............................ | 19 | 2 | 1.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTCGTGTCTGTTCCCCTGCAGA................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................CTGGGGGTACACAGTCGCG.............................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................GGGCCCGGCCTCCGCGCC | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ......................................................................................................................................................................................................AGAACCGGGCTGTGGGTCGGCCCACGGG........................ | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGAACCGGGCTGTGGGTCGGCAA............................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................AAGGGGAAGCTGGGCCCGGCCTCCGCGGCG | 30 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGAACCGGGCTGTGGGTCGG................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTTTCCGGGGGTCTGTCCC................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................GATCACACTCTGAGGACG............................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................TCCCCTGCAGAACCGAACT......................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................ACTCTGAGGAATGGGGCCA..................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................AACCGGGCTGTGGGTCGGC............................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................ATCACACTCTGAGGACC............................................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............ACCTCGTTTAACCTGACGGTCTCTGC.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAACCGGGCTGTGGGTCGGCC.............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CCGGGCTGTGGGTCGGC............................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...........................................................................................................................................................................................................CGGGCTGTGGGTCGGC............................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .........................................................................................................................................................................................TCTGTTCCCCTGCAGA................................................. | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| TTCACCTGAATTACCTCGTTTAACCTGACGGTCTCTGCAAAGCCCCCATCTCCAAATAAAGATCACACTCTGAGGAATGGGGTTTAGAGCCCCCATCGTGTCTTTCTGGGGGTACACAGTTTGCTCCCTAACACCCTTTCCGGGGGTCTGGCCGTGCCTGCAGTCTCCCCATCTGACCAGTCGTGTCTGTTCCCCTGCAGAACCGGGCTGTGGGTCGGCCCAAGGGGAAGCTGGGCCCGGCCTCCGCGGT ................................................................((((.(((.((((.(((((((((.(((((((...........)))))))....((....))..)))).))))))))).((((...(((.((....)).))).)))).......)))..))))((((......)))).................................................. ........................................................57.............................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR343334 | SRR343335 | SRR343336 | SRR343337 | SRR189784 | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR040007(GSM532892) G601T. (cervix) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR038857(GSM458540) D20. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | TAX577579(Rovira) total RNA. (breast) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM532876(GSM532876) G547T. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................CTCTGCAAAGCCCCCCGG........................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................CCTTTCCGGGGGTCTGTGT................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................ATTTGGAGATGGGGGCTTT................................................................................................................................................................................................. | 19 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |