| (1) AGO2.ip | (6) B-CELL | (3) BRAIN | (5) BREAST | (17) CELL-LINE | (2) CERVIX | (4) HEART | (1) HELA | (9) LIVER | (7) OTHER | (17) SKIN | (1) TESTES | (2) UTERUS | (1) XRN.ip |
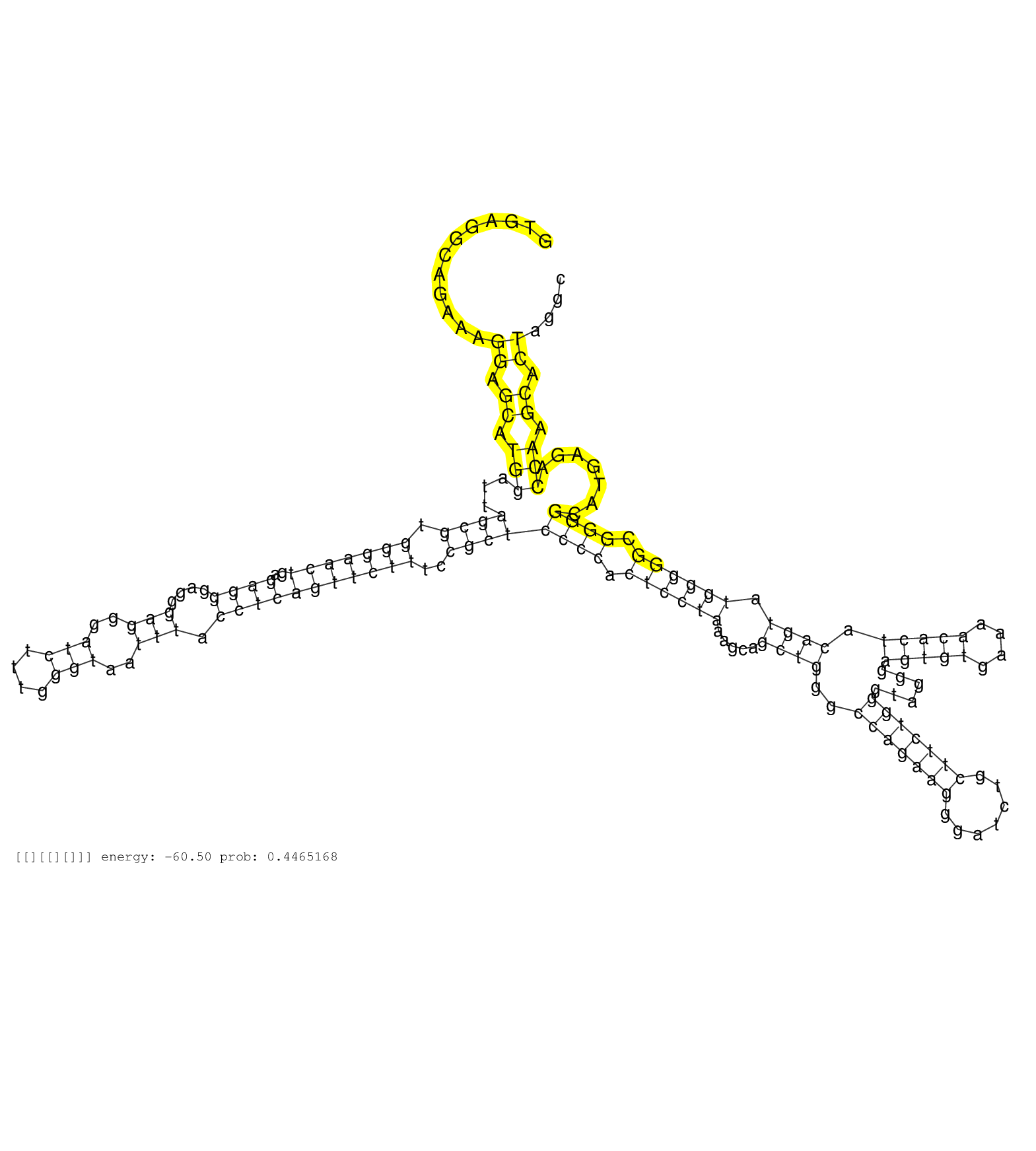
| TCATGAAGCTCTGGAGAAAGGTTCCAGCAGCTGACAAAGCCCCCTACCTGGTGAGGCAGAAAGGAGCATGGATTAGCGTGGGAACTGAGAGGGAGGGAGGGATCTTTGGGTAATTTACCTCAGTTCTTTCCGCTCCCCACTCCTAAAGCAGCTGGGCCAGAAGGGATCTGCTTCTGGGTAGGGAGTGTGAAAACACTACAGTATGGGGGCGGGGCATGAGACCAAGCACTAGGCCCTAAGGCTGTGTCCCATATCCCTCAGCAAAAGGCCAAAGATAACCGGGCAGCTCACCGCATCAACAAGGTGCAGAA ...........................................................................................................................................................................................................((((((.(((((((..............((((....)))))))))))....))))))................................................... ..........................................................................................................................................................................................................203.......................................................261................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR189786 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189784 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR343334 | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343335 | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR191499(GSM715609) 6genomic small RNA (size selected RNA from to. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR189782 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR040008(GSM532893) G727N. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR033728(GSM497073) MALT (MALT413). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM359177(GSM359177) hela_nucl_a. (hela) | SRR040015(GSM532900) G623T. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR343336 | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR444062(SRX128910) Sample 27_3cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR343337 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGCACT................................................................................. | 23 | 1 | 22.00 | 22.00 | 6.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............AGAAAGGTTCCAGCAGCTGAC.................................................................................................................................................................................................................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ACAAAGCCCCCTACCTG..................................................................................................................................................................................................................................................................... | 17 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GCGGGGCATGAGACCAAGCACT................................................................................. | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGCA................................................................................... | 21 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGCAC.................................................................................. | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGCACC................................................................................. | 23 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................AGGCCAAAGATAACCGGGCAGCTCACCGCA................ | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GATCTGCTTCTGGGTCGG................................................................................................................................. | 18 | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............AGAAAGGTTCCAGCAGCTGACA................................................................................................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................AACCGGGCAGCTCACCGCATC.............. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGCAAT................................................................................. | 23 | 1 | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AGCTCTGGAGAAAGGTGGCT............................................................................................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................................GCTGTGTCCCATATCCCTCAGT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................AGGTTCCAGCAGCTGACAA.................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................GATAACCGGGCAGCTCACCGCATCAACAAGGT...... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TATGGGGGCGGGGCATGAG........................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................GCTGGGCCAGAAGGGATCTGCT........................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................GGCCCTAAGGCTGTGTCCCATATCCCTCAG.................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................CCAAAGATAACCGGGCAGCT....................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGCACTC................................................................................ | 24 | 1 | 1.00 | 22.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ATGAAGCTCTGGAGAAAGGTTCCAGC........................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GGGAGGGAGGGATCTAAAA.......................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................TGGATTAGCGTGGGAACTGAGAGGGAGGGAGGG.................................................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGGGTAGGGAGTGTGAAAAC..................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAT..................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................TGAGACCAAGCACTAGACCT........................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................GCGGGGCATGAGACCAAGCAA.................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................AAAGGTTCCAGCAGCTGAC.................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAG..................................................................................... | 19 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAACA.................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGGGTAGGGAGTGTGTAAC...................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................................................................GATAACCGGGCAGCTCACCGCA................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CTGGTGAGGCAGAAAGGAGC.................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGC.................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................AACCGGGCAGCTCACAGCG................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................TGGTGAGGCAGAAAGGAGCATG................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGGATTAGCGTGGGATTG................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................................................................ATAACCGGGCAGCTCACCGCAT............... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AAAGCAGCTGGGCCAGAAGGGATCTG............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ATGAAGCTCTGGAGAAAGGT................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TATGGGGGCGGGGCATGAT........................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................TGCTTCTGGGTAGGGAGT............................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................GCAAAAGGCCAAAGATAACCG.............................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGCACTT................................................................................ | 24 | 1 | 1.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AGGTTCCAGCAGCTGACA................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................AACCGGGCAGCTCACCGCATAA............. | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............GAAAGGTTCCAGCAGCTGACA................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CTCTGGAGAAAGGTTCCAGCAGCTGACAAAGC............................................................................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGAAGCTCTGGAGAAAGG.................................................................................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGGAGAAAGGTTCCATG........................................................................................................................................................................................................................................................................................... | 17 | 8 | 0.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | 0.12 | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | 0.12 | - | - | - | 0.12 | 0.12 |
| ..............AGAAAGGTTCCAGCAG......................................................................................................................................................................................................................................................................................... | 16 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GCGGGGCATGAGACCA....................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................ACCGGGCAGCTCACC................... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GAGAAAGGTTCCAGCAGC........................................................................................................................................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGCGGGGCATGAGACCAAGAAA.................................................................................. | 22 | 2 | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................AAGGCCAAAGATAACC............................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGGAGAAAGGTTCCATGTG......................................................................................................................................................................................................................................................................................... | 19 | 8 | 0.25 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGGAGAAAGGTTCCATGGT......................................................................................................................................................................................................................................................................................... | 19 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - |
| ...........TGGAGAAAGGTTCCACGT.......................................................................................................................................................................................................................................................................................... | 18 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - |
| ...........TGGAGAAAGGTTCCATGT.......................................................................................................................................................................................................................................................................................... | 18 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - |
| ...........TGGAGAAAGGTTCCATGG.......................................................................................................................................................................................................................................................................................... | 18 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - |
| ...........TGGAGAAAGGTTCCA............................................................................................................................................................................................................................................................................................. | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - |
| TCATGAAGCTCTGGAGAAAGGTTCCAGCAGCTGACAAAGCCCCCTACCTGGTGAGGCAGAAAGGAGCATGGATTAGCGTGGGAACTGAGAGGGAGGGAGGGATCTTTGGGTAATTTACCTCAGTTCTTTCCGCTCCCCACTCCTAAAGCAGCTGGGCCAGAAGGGATCTGCTTCTGGGTAGGGAGTGTGAAAACACTACAGTATGGGGGCGGGGCATGAGACCAAGCACTAGGCCCTAAGGCTGTGTCCCATATCCCTCAGCAAAAGGCCAAAGATAACCGGGCAGCTCACCGCATCAACAAGGTGCAGAA ...........................................................................................................................................................................................................((((((.(((((((..............((((....)))))))))))....))))))................................................... ..........................................................................................................................................................................................................203.......................................................261................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR189786 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189784 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR343334 | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343335 | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR191499(GSM715609) 6genomic small RNA (size selected RNA from to. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR189782 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR040008(GSM532893) G727N. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR033728(GSM497073) MALT (MALT413). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM359177(GSM359177) hela_nucl_a. (hela) | SRR040015(GSM532900) G623T. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR343336 | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR444062(SRX128910) Sample 27_3cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR343337 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................GTAGGGAGTGTGAAAAG..................................................................................................................... | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................CGCTCCCCACTCCTAAA.................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................ATGGGGGCGGGGCATGATG.......................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................AGGTTCCAGCAGCTGCTG................................................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................CAAAGATAACCGGGCT.......................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................TGAGACCAAGCACTACCT............................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |