| (2) AGO2.ip | (3) B-CELL | (4) BREAST | (12) CELL-LINE | (8) HEART | (1) KIDNEY | (2) LIVER | (2) OTHER | (17) SKIN | (1) TESTES |
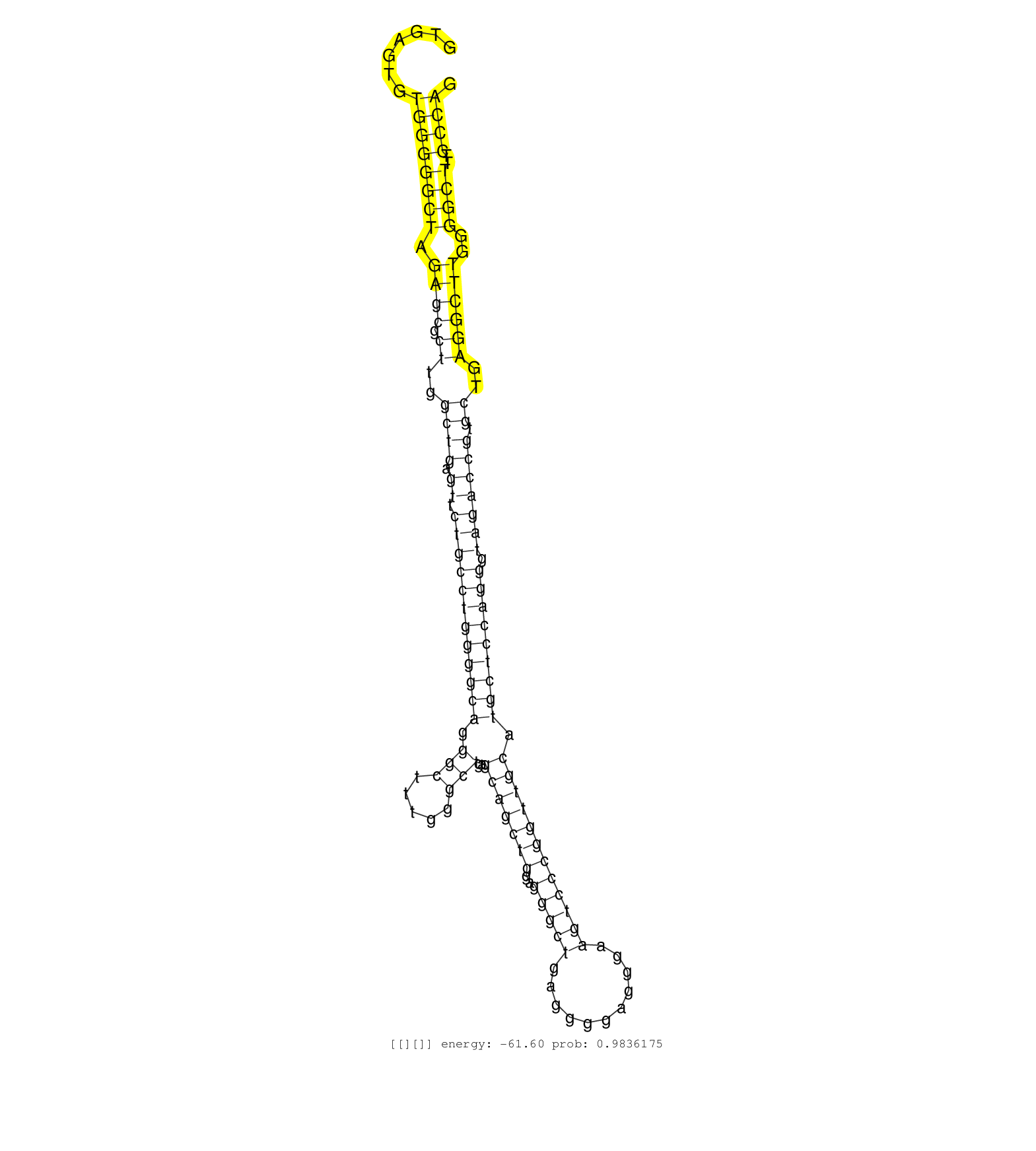
| GCCACTCGGGTCCCCAGAGTTCAGGAGGTGCTGCTGAAGGCCGTGGGCAAGTGAGTGTGGGGGCTAGAGCGCTTGGCTGAGTTCTGCCTGGGGCAGGGCTTTGGGCTGAGCAGCTGGAGGGCTGAGGGGAGGGAAGTCCCGGTTGCATGCTCCAGGGTAGACCGTGCTGAGGCTTGGGGCTTTCCCAGGGAGGAGCTGGGGAAGAACATCAATGCAGATGAAGCAGCCGCCATGGGGG .......................................................................................((((((..((((((..((((...((((.((((.((....((((......))))....))...)))).((.....))..)))).))))..)))))))))))).................................................. ....................................................................................85.....................................................................................................188................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR189784 | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189782 | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR029129(GSM416758) SW480. (cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR191471(GSM715581) 119genomic small RNA (size selected RNA from . (breast) | TAX577738(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCAGT................................................. | 22 | 1 | 24.00 | 1.00 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCAGA................................................. | 22 | 1 | 8.00 | 1.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGGGGAAGAACATCAATGCAGATGAAGCAGC........... | 31 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGCTGAGGCTTGGGGCTTTCCCAG.................................................. | 24 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CAGGAGGTGCTGCTGAAGG...................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCAGTAAT.............................................. | 25 | 1 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCAGTAT............................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CAGAGTTCAGGAGGTGCTGCTGAA........................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGGAGGTGCTGCTGAAGGCCGTG................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAAGAACATCAATGCAG..................... | 17 | 3 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAAGAACATCAATGCAGATGAAGCAGCCGCCAT..... | 33 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TTCAGGAGGTGCTGCTGA......................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........CCCCAGAGTTCAGGAGGTGCTGCTGAAGGCCGTG................................................................................................................................................................................................. | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GGGGCAGGGCTTTGGGCCCG................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................CATCAATGCAGATGAAGCAGCCGCCATGGG.. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AGGGAAGTCCCGGTTGCATGCT....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCAGGAA............................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GGAAGAACATCAATGCAGATGAAGCAGCCG......... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAAGAACATCAATGCAGATGAAGCAG............ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGTGCTGCTGAAGGCCGTGGGCAAG........................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................CCGTGGGCAAGTGAGTGTGGGGGC.............................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................GGGAGGAGCTGGGGATTTT................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................CTGCTGAAGGCCGTGGGC.............................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................AGCTGGGGAAGAACATCAATGCAGATGAAGCAG............ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGCTGAGGCTTGGGGCTTTCCCAGT................................................. | 25 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGGGGAAGAACATCAATGCAGATGAAGCAG............ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGTGGGGGCTAGAGTAG...................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................CAGGGAGGAGCTGGGGAAGAACACTG........................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................TGGGGAAGAACATCAATGCAGA.................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGGGGGCTAGA.......................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................GCTGGGGAAGAACATCAATGCA...................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AGGGAAGTCCCGGTTGCATGCTCT..................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................TGGGCTGAGCAGCTGGAGGGC.................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................AGGGAGGAGCTGGGGAAGAACACTG........................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................AGGAGGTGCTGCTGAAGG...................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AATGCAGATGAAGCAGCCGCCATGGGGG | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................GAGCTGGGGAAGAACATCAATGC....................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................GGGGAAGAACATCAATGCAGATGAAGCAG............ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TGAAGGCCGTGGGCAAG........................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................AGCTGGGGAAGAACATCAATG........................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CAGGAGGTGCTGCTGTAA....................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................GAAGTCCCGGTTGCATT......................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................CGTGGGCAAGTGAGTGTGGGGGC.............................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................AGGAGCTGGGGAAGAACATCAATGCAGATGAAG............... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................GAGGAGCTGGGGAAGATGG.............................. | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCAGTAGA.............................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGGCTTGGGGCTTTCCCATT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................TGGGGAAGAACATCAATGCAGATGAAGCAGT........... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAAGAACATCAATG........................ | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - |
| .............................................................................................................................................................................TTGGGGCTTTCCCAGG................................................. | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - |
| GCCACTCGGGTCCCCAGAGTTCAGGAGGTGCTGCTGAAGGCCGTGGGCAAGTGAGTGTGGGGGCTAGAGCGCTTGGCTGAGTTCTGCCTGGGGCAGGGCTTTGGGCTGAGCAGCTGGAGGGCTGAGGGGAGGGAAGTCCCGGTTGCATGCTCCAGGGTAGACCGTGCTGAGGCTTGGGGCTTTCCCAGGGAGGAGCTGGGGAAGAACATCAATGCAGATGAAGCAGCCGCCATGGGGG .......................................................................................((((((..((((((..((((...((((.((((.((....((((......))))....))...)))).((.....))..)))).))))..)))))))))))).................................................. ....................................................................................85.....................................................................................................188................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR189784 | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189782 | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR029129(GSM416758) SW480. (cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR191471(GSM715581) 119genomic small RNA (size selected RNA from . (breast) | TAX577738(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............CACCTCCTGAACTCTG................................................................................................................................................................................................................ | 16 | 3 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | 0.33 | 1.33 | 0.33 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | - | - | - | - |
| ..............ACCTCCTGAACTCTG................................................................................................................................................................................................................. | 15 | 9 | 1.22 | 1.22 | - | - | - | - | - | - | - | 0.11 | - | - | 0.11 | 0.11 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | 0.11 | - | - | - | - | - | - | - | - | 0.11 | - | 0.11 | 0.11 |
| ..............GCAGCACCTCCTGAACTCTG............................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CATGCTCCAGGGTAGTTT........................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............GCACCTCCTGAACTCTG............................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............AGCACCTCCTGAACTCTG.............................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |