| (1) BRAIN | (1) BREAST | (13) CELL-LINE | (2) HEART | (4) HELA | (1) KIDNEY | (2) LIVER | (1) OTHER | (7) SKIN | (1) TESTES | (2) XRN.ip |
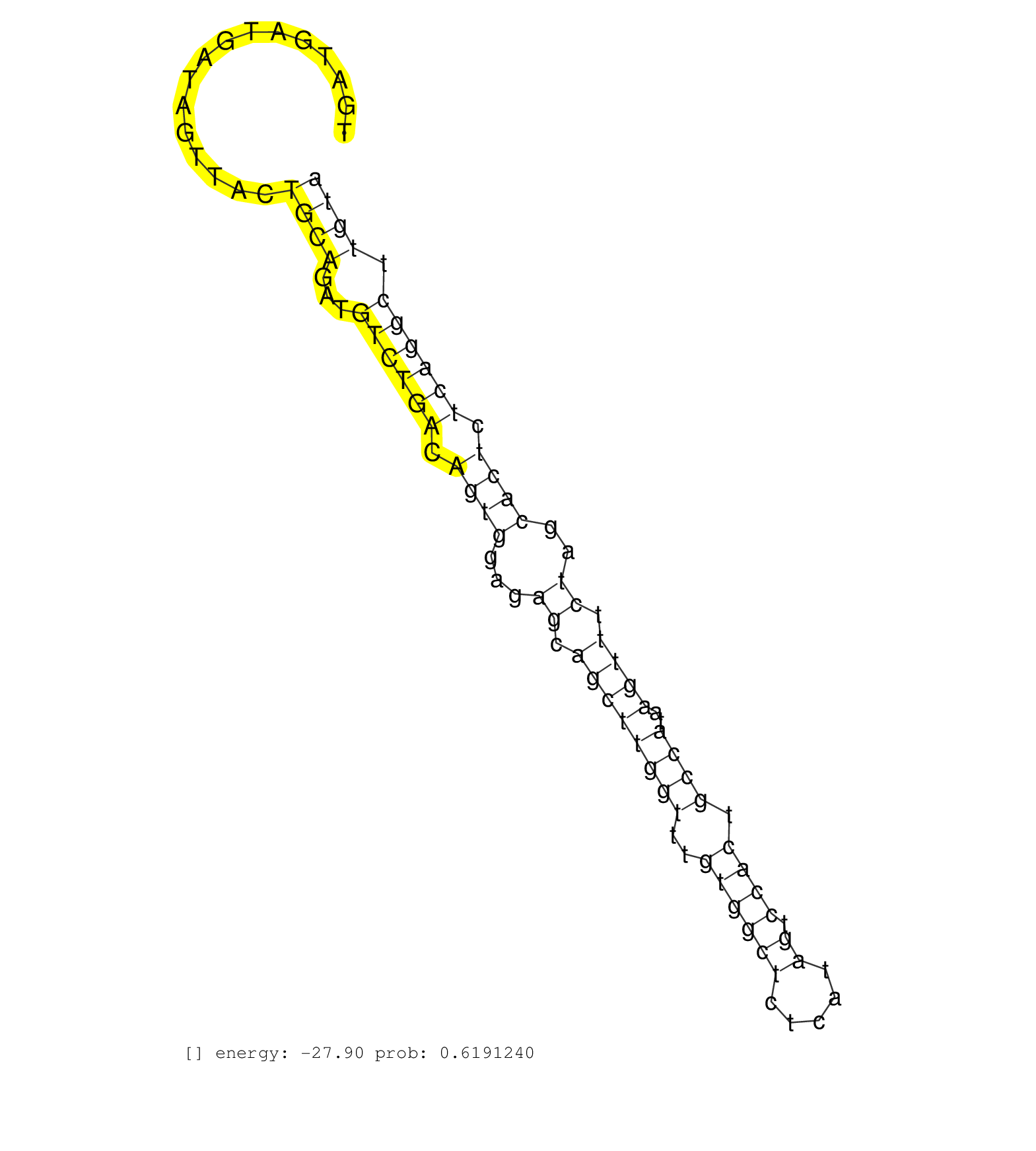
| GAGAAGTTGAAAGTGCAAAAAAGCAGTTAGAAGAAACACAACACGATAAGGTACTGAAATCTTCTCTAAATCCATGAAGAGCCAAGTTGAACTGAGTTGATGATGATGATAGTTACTGCAGATGTCTGACAGTGGAGAGCAGCTTGGTTTGTGGCTCTCATAGTCCACTGCCATAAGTTTCTAGCACTCTCAGGCTTGTATGTTGAATGCCTTTGTATTTCAGGTGATTTTGAGTCTGGCTATGCATATG ...............................................................((((..........))))....((..(((((.(((......)))..)))))...))................................................................................................................................... ...............................................................64............................................................126.......................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553575(SRX182781) source: Kidney. (Kidney) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR189784 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR553576(SRX182782) source: Testis. (testes) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR343337 | TAX577743(Rovira) total RNA. (breast) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................GATGATGATGATAGTTACTGCAGATGTCTGACA....................................................................................................................... | 33 | 1 | 99.00 | 99.00 | 76.00 | 9.00 | 6.00 | 1.00 | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................ATGATGATGATAGTTACTGCAGATGTCTGACA....................................................................................................................... | 32 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................GCAGATGTCTGACAGTGGAGAGCAGCT.......................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GATGATGATGATAGTTACTGCAGATGTCTG.......................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TCAGGCTTGTATGTTGAATGA........................................ | 21 | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................GATGATGATGATAGTTACTGCAGATGTCTGAC........................................................................................................................ | 32 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGATGATGATGATAGTTACTGCAGATGTCTGACA....................................................................................................................... | 34 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGATGATAGTTACTGCAGA................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................ATCCATGAAGAGCCAAGTTGAACTGAGTTGATGATG................................................................................................................................................. | 36 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................ATCCATGAAGAGCCAAGTTGAACTGAGTTGATGATGATGATAGTTACTGCAGATG.............................................................................................................................. | 55 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................AATCCATGAAGAGCCCTTA................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................TGATAGTTACTGCAGATGTCTGACA....................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GATGATGATGATAGTTACTGCAG................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................TGATGATAGTTACTGCAGATGTCTGA......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................ATGTCTGACAGTGGAGAGC.............................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................ATGTCTGACAGTGGAGAGCAGCTTGGTCTG................................................................................................... | 30 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................AAAAAGCAGTTAGAAGAAACATA.................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................AATCCATGAAGAGCCAAG.................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GATGATGATGATAGTTACTGCA.................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AAGCAGTTAGAAGAAACAC................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................ACACAACACGATAAGGTACTGAAATC............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................CATGAAGAGCCAAGTTGAACTGAGTTGATGA................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CTGCAGATGTCTGACAGTGGAGAGCAG............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........AGTGCAAAAAAGCAGTTAGAAGAAACAC................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TGAGTCTGGCTATGCATATG | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................ATCCATGAAGAGCCAAGTTGAACTG............................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGCTTGGTTTGTGGCTCGTA.......................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ..................................................................................................GATGATGATGATAGTTACTGCAGA................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AAAAGCAGTTAGAAGAAACACA.................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TCAGGTGATTTTGAGTCTGA........... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................ATCCATGAAGAGCCAAGTTGAACTGAGT......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AAAAGCAGTTAGAAGAAACAC................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................AAAAAGCAGTTAGAAGAAACACA.................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........AAGTGCAAAAAAGCAGTAAGA........................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ................................................................................................................TTACTGCAGATGTCTGACA....................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CCAAGTTGAACTGAG.......................................................................................................................................................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| GAGAAGTTGAAAGTGCAAAAAAGCAGTTAGAAGAAACACAACACGATAAGGTACTGAAATCTTCTCTAAATCCATGAAGAGCCAAGTTGAACTGAGTTGATGATGATGATAGTTACTGCAGATGTCTGACAGTGGAGAGCAGCTTGGTTTGTGGCTCTCATAGTCCACTGCCATAAGTTTCTAGCACTCTCAGGCTTGTATGTTGAATGCCTTTGTATTTCAGGTGATTTTGAGTCTGGCTATGCATATG ...............................................................((((..........))))....((..(((((.(((......)))..)))))...))................................................................................................................................... ...............................................................64............................................................126.......................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553575(SRX182781) source: Kidney. (Kidney) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR189784 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR553576(SRX182782) source: Testis. (testes) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR343337 | TAX577743(Rovira) total RNA. (breast) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................TAAGGTACTGAAATCCAC.......................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................GTTAGAAGAAACACAACCTG............................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................ATAAGGTACTGAAATCAC........................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....AGTTGAAAGTGCAAAAGTG................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................TGATGATGATGATAGTCAAA..................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................ATGGATTTAGAGAAGATT............................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |