| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (3) AGO2.ip | (1) AGO3.ip | (1) B-CELL | (1) BRAIN | (6) BREAST | (19) CELL-LINE | (1) HEART | (4) HELA | (1) LIVER | (1) OTHER | (9) SKIN | (1) XRN.ip |
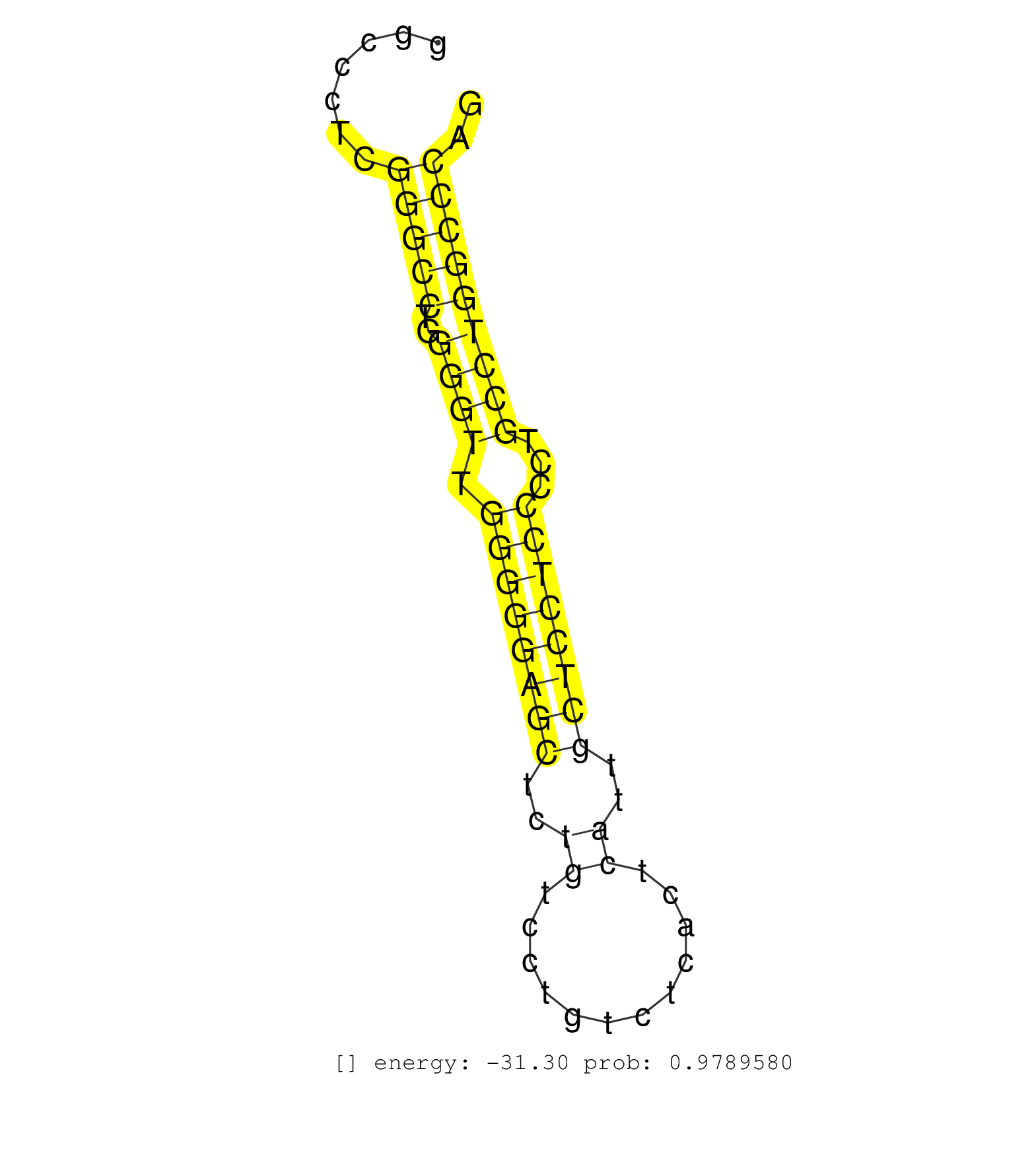
| GTCCACATCTTTCTGCCCAGCAAGGAGGTCTACGAGAGCATCTACAACAGGTGGAGACCCGGGGCCAGCAGGGCGGGACTGGACACTGGGTGGGCCTGGCCTCCCTTACCTGCTGTCCACCCGCCTATCTCAGGATCAACAACGACCTGCTCATGTGGGAGCCTGCAGATCTGCTTCCCACCC ......................................................................(((((((..((((((..(((((((........)))..))))..)))))))))))))......................................................... ...............................................................64...................................................................133................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | TAX577588(Rovira) total RNA. (breast) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR191574(GSM715684) 78genomic small RNA (size selected RNA from t. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191633(GSM715743) 85genomic small RNA (size selected RNA from t. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | TAX577743(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................CAGGGCGGGACTGGACACTGG.............................................................................................. | 21 | 1 | 28.00 | 28.00 | - | 5.00 | 12.00 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGGT............................................................................................ | 23 | 1 | 26.00 | 26.00 | 13.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTGG.......................................................................................... | 24 | 1 | 9.00 | 9.00 | 3.00 | - | - | 1.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACT................................................................................................ | 19 | 1 | 8.00 | 8.00 | - | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGT............................................................................................ | 22 | 1 | 8.00 | 8.00 | 1.00 | 2.00 | - | - | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGG.............................................................................................. | 20 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGA............................................................................................. | 22 | 1 | 5.00 | 28.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTGT.......................................................................................... | 24 | 4.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | |
| ....................................................................CAGGGCGGGACTGGACACTGGG............................................................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTA........................................................................................... | 23 | 1 | 3.00 | 8.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGAA............................................................................................ | 23 | 1 | 3.00 | 28.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGGTGGA......................................................................................... | 26 | 3.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................CAGGGCGGGACTGGACACTGT.............................................................................................. | 21 | 1 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGAGACCCGGGGCCAGCAGGGCGGGACTGGACACTGGGTGGGCCTGGCCTCCCTTACCTGCTGTCCACCCGCCTATCTCAG.................................................. | 83 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGGTTT.......................................................................................... | 25 | 1 | 3.00 | 26.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGATC........................................................................................... | 24 | 1 | 3.00 | 28.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGGTGTA......................................................................................... | 26 | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................AGGGCGGGACTGGACACTGGG............................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGT.............................................................................................. | 20 | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................GCAGGGCGGGACTGGACACTGT.............................................................................................. | 22 | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................CAGGGCGGGACTGGACACTGGGA............................................................................................ | 23 | 1 | 2.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GCAGGGCGGGACTGGACACTGGG............................................................................................. | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGT............................................................................................. | 22 | 1 | 2.00 | 28.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................CTGCTGTCCACCCGCCTATCT..................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTGGA......................................................................................... | 25 | 1 | 2.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CATGTGGGAGCCTGCAGAT............. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................AGGAGGTCTACGAGAGCATCTACA......................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGG............................................................................................ | 22 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTAGA......................................................................................... | 25 | 1 | 1.00 | 8.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTAGGT............................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................CAGGGCGGGACTGGACACTGTTT............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTGTTT........................................................................................ | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................ACCTGCTGTCCACCCGCCTATC...................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGGAA........................................................................................... | 24 | 1 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGGGCA.......................................................................................... | 25 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTGA.......................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................CAGGGCGGGACTGGACACTGAAA............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGTAA............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACACTGGGTTG.......................................................................................... | 24 | 1 | 1.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGATGT.......................................................................................... | 25 | 1 | 1.00 | 28.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GCAGGGCGGGACTGGACACTGGT............................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................ACTGGACACTGGGTGCTTG....................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................CTGCTGTCCACCCGCCTATCTCTT.................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................CAGGGCGGGACTGGACACTG............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................ACCTGCTGTCCACCCGCCTATCT..................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................AGCAGGGCGGGACTGGACACTGA.............................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................CAACGACCTGCTCATGTGGGAGCCTGCAGA.............. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGGGCGGGACTGGACACTGGGTGGTTT....................................................................................... | 28 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................AGGGCGGGACTGGACACCGGA............................................................................................. | 21 | 2 | 1.00 | 0.50 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GAGCCTGCAGATCTGCTTCCCA... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGCGGGACTGGACAC................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTCCACATCTTTCTGCCCAGCAAGGAGGTCTACGAGAGCATCTACAACAGGTGGAGACCCGGGGCCAGCAGGGCGGGACTGGACACTGGGTGGGCCTGGCCTCCCTTACCTGCTGTCCACCCGCCTATCTCAGGATCAACAACGACCTGCTCATGTGGGAGCCTGCAGATCTGCTTCCCACCC ......................................................................(((((((..((((((..(((((((........)))..))))..)))))))))))))......................................................... ...............................................................64...................................................................133................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | TAX577588(Rovira) total RNA. (breast) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR191574(GSM715684) 78genomic small RNA (size selected RNA from t. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191633(GSM715743) 85genomic small RNA (size selected RNA from t. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | TAX577743(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................AGATCTGCTTCCCACCAA | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................CCCGGGGCCAGCAGGGCGTC.......................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |