| (1) AGO1.ip OTHER.mut | (1) AGO2.ip | (11) B-CELL | (3) BRAIN | (9) BREAST | (20) CELL-LINE | (3) CERVIX | (2) HEART | (2) LIVER | (2) OTHER | (1) OVARY | (10) SKIN |
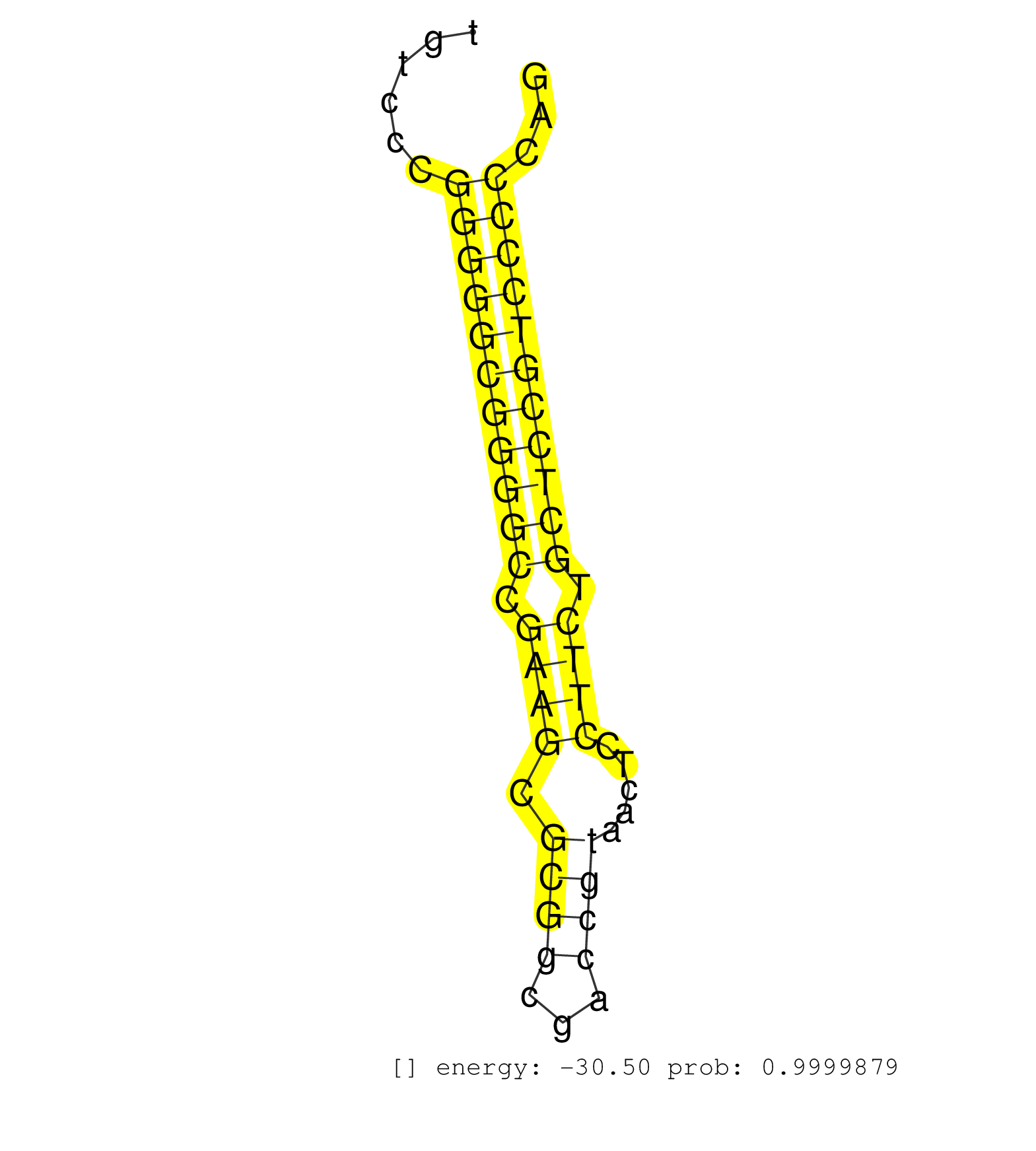
| CTGGAGACCGGCCAGGTCGGGCAGCGGTGGCCAGGAGCGCTATGATGCAGGTGGGCTGGGCTGGGCTGGGTTGGGGGGAAATTGGTTTTAGGAGAGGCCTAGGCCCGGGGACTCTAGGCCTAGATCCAACTGGCGCCCGCAGCGTGAGCTCAGCAAGGTCCTAAGCCTCTCCGACATGGGGCGTGGCGGGGCCGCGGGGCTACCAGGTGGGACCTCTGACGCGCCGCCTTCGCCTTCGCCTTCGCCTTCGCCTCCAGGACTCGCCCTTCTTCCAGCAGTACGAGCTGGACCTGCGGGAGCCTGCGCTGGGCCAGGGCAGCTTTTC ....................................................(((((..((((((((..((((.(((....((((((...(((.((((((((.((...))...))))))))...)))))))))..))).))))...))))))))..)))))...(((((.((.((.......)))).)))))..................................................................................................................................... ..................................................51............................................................................................................................................193.................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | TAX577745(Rovira) total RNA. (breast) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | TAX577580(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR189785 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR040007(GSM532892) G601T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577741(Rovira) total RNA. (breast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | GSM379265(GSM379265) Small RNA from endometrioid ovarian cancer tissue. (ovary) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR191455(GSM715565) 181genomic small RNA (size selected RNA from . (breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR040041(GSM532926) G612T. (cervix) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................CTGGGCTGGGCTGGGTTGGGCA........................................................................................................................................................................................................................................................ | 22 | 7.00 | 0.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................CTGGGCTGGGCTGGGTTGGGCAA....................................................................................................................................................................................................................................................... | 23 | 6.00 | 0.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGGTTTTT.......................................................................................................................................................................................................................................................... | 25 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................CTGGGCTGGGTTGGGCAA....................................................................................................................................................................................................................................................... | 18 | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGC................................................................................................................................................................................................................................................................... | 16 | 0 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CTGGGCTGGGCTGGGTAGG........................................................................................................................................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................CTGGGCTGGGCTGGGTTGGGGGA....................................................................................................................................................................................................................................................... | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGTCC............................................................................................................................................................................................................................................................. | 22 | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CGGCCAGGTCGGGCAGCGGTGGCCAGGAGCG.............................................................................................................................................................................................................................................................................................. | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CTGGGCTGGGCTGGGTTGGGCAG....................................................................................................................................................................................................................................................... | 23 | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................AGGAGCGCTATGATGCAG................................................................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGCTGGGCTGGGA.............................................................................................................................................................................................................................................................. | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................................................................................GAGCTGGACCTGCGGGAGCC........................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TCGGGCAGCGGTGGCCAGGAGC............................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............GTCGGGCAGCGGTGGCCAGGA................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGGAGACCGGCCAGGTCGGGCAGC............................................................................................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGCTGGGCTGTGG.............................................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................CTGGGCTGGGCTGGGTTGAGCC........................................................................................................................................................................................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTAGGC.............................................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGGGA............................................................................................................................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................................................................................AGCTGGACCTGCGGGAAGAC....................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................CTGGGCTGGGTTGGGCATT...................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................CTGGGCTGGGCTGGGTTGGGCCA....................................................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGGATA............................................................................................................................................................................................................................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................................................................................CGGGAGCCTGCGCTGGGCCAGGGCAGCT.... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................CAGTACGAGCTGGACCTGCGGGAG.......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGCTGGGCTGGGAG............................................................................................................................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGGAGAA........................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................................................................................................AGCCTGCGCTGGGCCAGGG......... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGCTGGGCTGGTCG............................................................................................................................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................CTGGGCTGGGCTGGGTTGGGCTA....................................................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGACCA............................................................................................................................................................................................................................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................GCTGGGCTGGGTTGGGCAA....................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGTGC.............................................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................................................................................AGCTGGACCTGCGGGAGCCTG...................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GGTCGGGCAGCGGTGGTCGG................................................................................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................................................................................................CTGCGCTGGGCCAGGGCAGCTTTTC | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGCTGGGCTGTG............................................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGGGGCGTGGCGGGGTTC................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................CAGGACTCGCCCTTCTTCCAGCAGTACGAGCT....................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................CGGGCAGCGGTGGCCA.................................................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................AGCAGTACGAGCTGGACCTGCGG............................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................TCTTCCAGCAGTACGAGC........................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GCCAGGTCGGGCAGCGGTGGCCAGGA................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGCTGGGCTGGGTAC............................................................................................................................................................................................................................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGGAAAG........................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................TGGGCTGGGCTGGGTTGGGCAA....................................................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGGTAA............................................................................................................................................................................................................................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................TCGGGCAGCGGTGGCCAGGAG................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGCTGGGCTGGTCAA............................................................................................................................................................................................................................................................ | 23 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGGGCTGGGCTGGGCTGGGGATT........................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................TGGACCTGCGGGAGC......................... | 15 | 5 | 0.80 | 0.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | 0.20 | 0.20 | - | - |
| ...........................................................................................................................................................................................................................................................................................GCTGGACCTGCGGGAGC......................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ................................................AGGTGGGCTGGGCTGGGCTGG................................................................................................................................................................................................................................................................ | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................TGCGCTGGGCCAGGGCA....... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................TGCGCTGGGCCAGGGC........ | 16 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGGGCTGGGCTGGGCTGGGTT............................................................................................................................................................................................................................................................. | 21 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| CTGGAGACCGGCCAGGTCGGGCAGCGGTGGCCAGGAGCGCTATGATGCAGGTGGGCTGGGCTGGGCTGGGTTGGGGGGAAATTGGTTTTAGGAGAGGCCTAGGCCCGGGGACTCTAGGCCTAGATCCAACTGGCGCCCGCAGCGTGAGCTCAGCAAGGTCCTAAGCCTCTCCGACATGGGGCGTGGCGGGGCCGCGGGGCTACCAGGTGGGACCTCTGACGCGCCGCCTTCGCCTTCGCCTTCGCCTTCGCCTCCAGGACTCGCCCTTCTTCCAGCAGTACGAGCTGGACCTGCGGGAGCCTGCGCTGGGCCAGGGCAGCTTTTC ....................................................(((((..((((((((..((((.(((....((((((...(((.((((((((.((...))...))))))))...)))))))))..))).))))...))))))))..)))))...(((((.((.((.......)))).)))))..................................................................................................................................... ..................................................51............................................................................................................................................193.................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | TAX577745(Rovira) total RNA. (breast) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | TAX577580(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR189785 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR040007(GSM532892) G601T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577741(Rovira) total RNA. (breast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | GSM379265(GSM379265) Small RNA from endometrioid ovarian cancer tissue. (ovary) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR191455(GSM715565) 181genomic small RNA (size selected RNA from . (breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR040041(GSM532926) G612T. (cervix) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................AGGCGAAGGCGAAGGCGG.................................................................................... | 18 | 2 | 2.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | 0.50 | - | - | - | - | - |
| ........................................................................................................................................................................................GGCGGGGCCGCGGGGGGGG.......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................CCCAGCCCAGCCCACCTGCAT.................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GAAGGCGAAGGCGAAGGCGG.................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................AGATCCAACTGGCGCCTCTT........................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................GAGCTCAGCAAGGTCCTAAGTG.............................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................AAGGCGAAGGCGAAGGCG................................................................................... | 18 | 5 | 0.80 | 0.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................AGGCGAAGGCGAAGGCGGC.................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AAGGCGAAGGCGAAGGCGG................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| .................................................................TTTCCCCCCAACCCAG.................................................................................................................................................................................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |