| (1) BRAIN | (1) BREAST | (3) CELL-LINE | (1) LIVER | (1) OTHER | (2) SKIN |
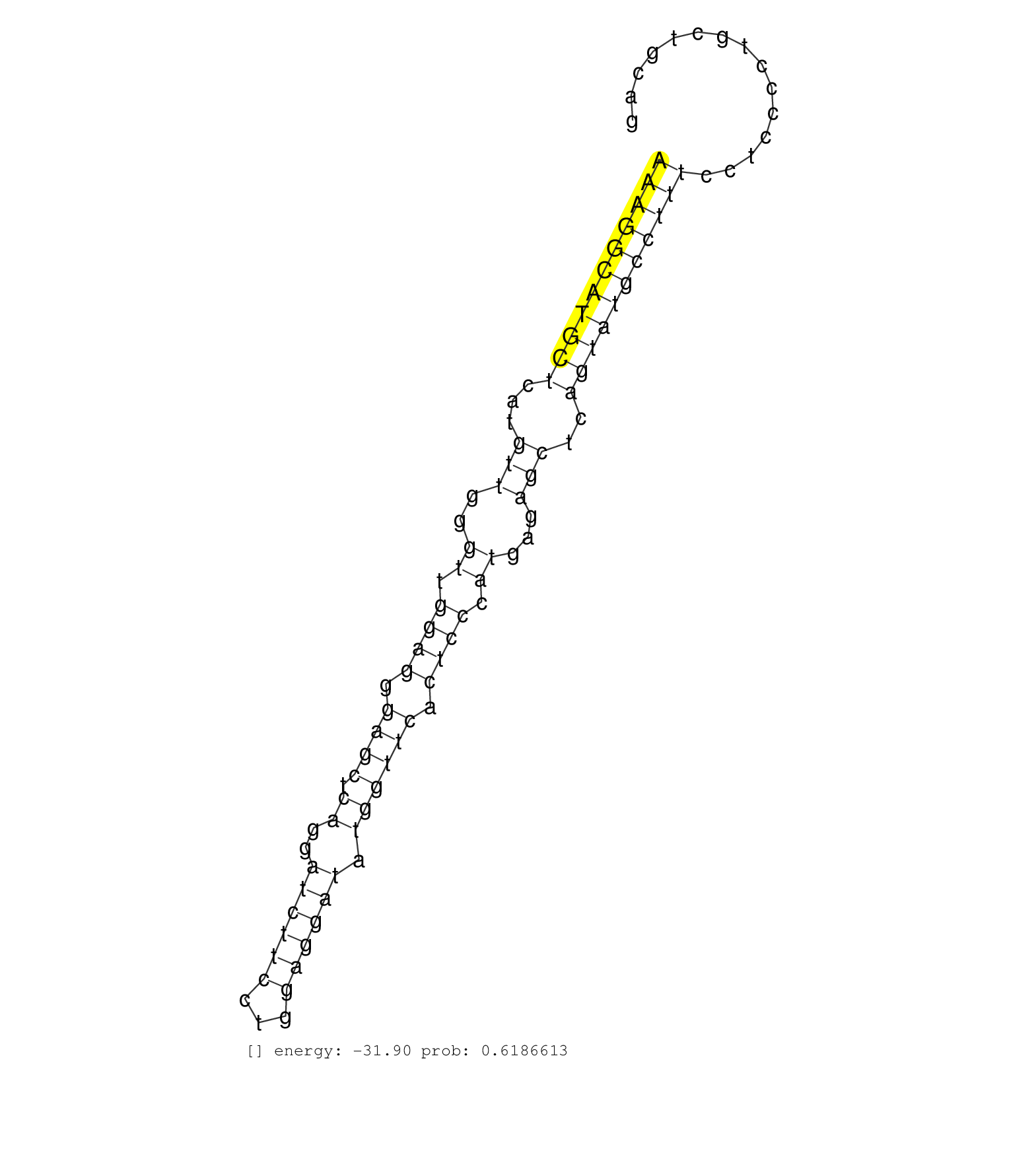
| GCAACCTTCCCTGTAGTTAGAAAAACGTTTAGTTGGATTTATCAGTCTGATTTTTCACTCATAAACAATTGCACAAGCCTTTGCAAGCCCAGAGGGTCTTGTAATAATGATGGCCGTGGTCCATTATGGCACAGTGATGTGTTCTACGGTGGTCATTTTGCTGGCAATGACAGTCACCTCTGAACTTCTCTTTCCAACAGACACAATTTCTCGGGGTTTACTTTGTCTCATCCCAAGAGTTCCTTGCCAA ....................................................................................(((..(((((((((((((...((((((.(((((((........((((((....))))))))))))).))))))......)))).))).....))))))..)))............................................................... ...................................................................................84...........................................................................................................193....................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................CCAGAGGGTCTTGTAGGG................................................................................................................................................ | 18 | 89.00 | 0.00 | 89.00 | - | - | - | - | - | - | - | - | - | |
| .......................................................................................CCCAGAGGGTCTTGTAGGG................................................................................................................................................ | 19 | 6.00 | 0.00 | 6.00 | - | - | - | - | - | - | - | - | - | |
| .................TAGAAAAACGTTTAGGATC...................................................................................................................................................................................................................... | 19 | 4.00 | 0.00 | - | 4.00 | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................AATTTCTCGGGGTTTACTTAG......................... | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | |
| ......................................TTATCAGTCTGATTTTGAC................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | |
| ..........................................................................................AGAGGGTCTTGTAATACA.............................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | |
| ........................................................................................CCAGAGGGTCTTGTAGGGA............................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................GGCAATGACAGTCACCTCTGAACTTC.............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................AGTCACCTCTGAACTCTG............................................................. | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | |
| ..........................................................................................................................................................................CAGTCACCTCTGAACTCTG............................................................. | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | |
| .................................................................................................................CCGTGGTCCATTATG.......................................................................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| GCAACCTTCCCTGTAGTTAGAAAAACGTTTAGTTGGATTTATCAGTCTGATTTTTCACTCATAAACAATTGCACAAGCCTTTGCAAGCCCAGAGGGTCTTGTAATAATGATGGCCGTGGTCCATTATGGCACAGTGATGTGTTCTACGGTGGTCATTTTGCTGGCAATGACAGTCACCTCTGAACTTCTCTTTCCAACAGACACAATTTCTCGGGGTTTACTTTGTCTCATCCCAAGAGTTCCTTGCCAA ....................................................................................(((..(((((((((((((...((((((.(((((((........((((((....))))))))))))).))))))......)))).))).....))))))..)))............................................................... ...................................................................................84...........................................................................................................193....................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................AGAAGTTCAGAGGTGACTGT............................................................. | 20 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 |
| ...........................................................................................................................................................................AGAAGTTCAGAGGTGACT............................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 |