| (1) AGO2.ip | (1) BRAIN | (3) BREAST | (28) CELL-LINE | (1) FIBROBLAST | (1) HEART | (6) HELA | (3) LIVER | (1) OTHER | (1) RRP40.ip | (2) SKIN | (1) TESTES | (1) XRN.ip |
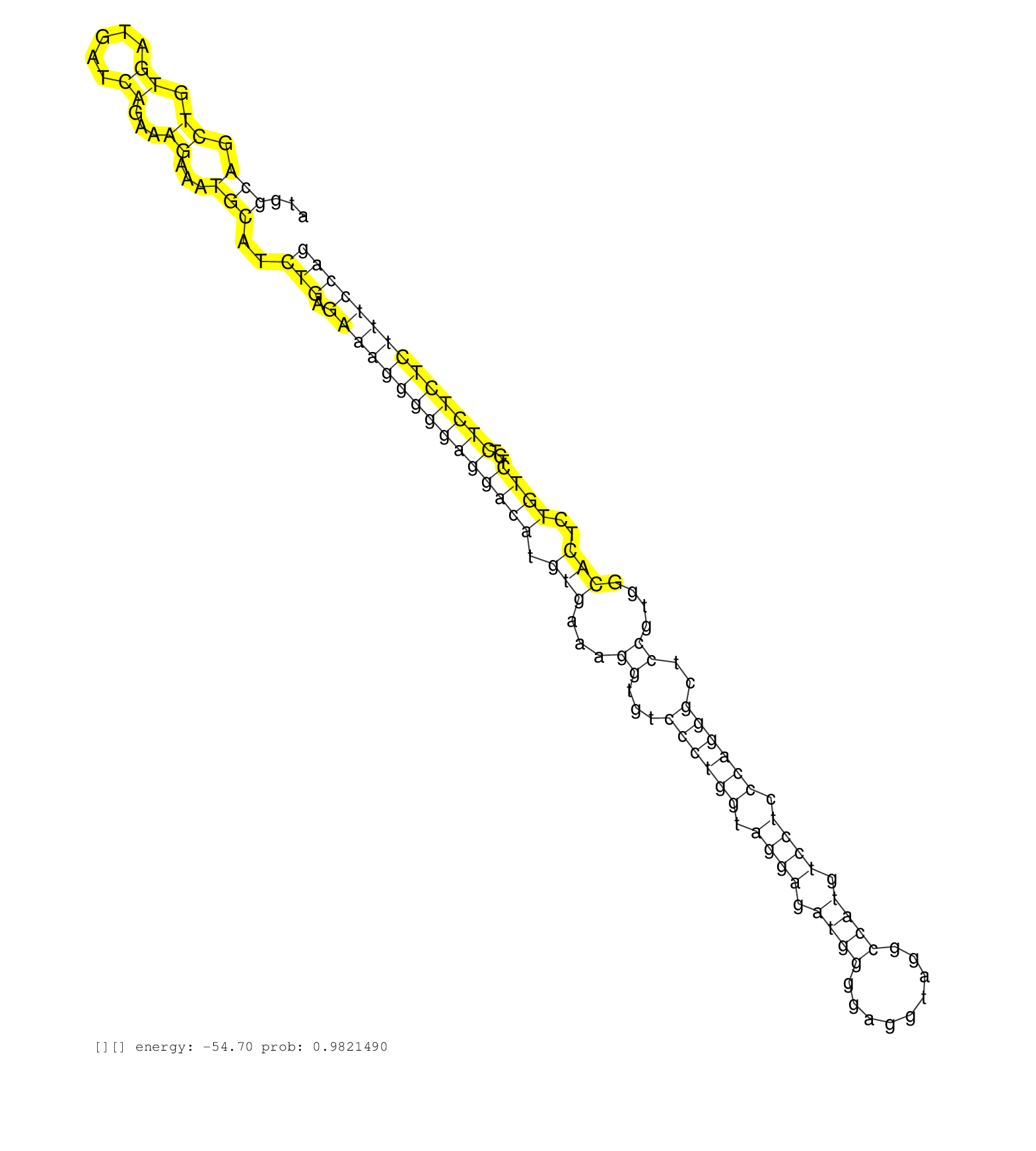
| TCTCTGAGGAGCCGAATTCCCTCCTCTGTCAAATAGGAATAACCCTTCTGTGTCTACCTCCCATGGCAGCTGTGATGATCAGAAAGAAATGCATCTGAGAAAGGGGGAGGACATGTGAAAGGTGTCCCTGGTAGGAGATGGGGAGGTAGGCCATGTCCTCCCAGGGCTCCGTGGCACTCTGTCTCTCTCTCTCTTTCCAGGAGTCCGCTCAGAATTTGATCAGATCGACACATCCAACCCAAACTGTGTG .................................................................(((.((.((.....))...))...)))..(((.(((((((((((((((.(((...((...((((((.((((.((((.........)))).)))).))))))..))....)))..))))...)))))))))))))).................................................. ..............................................................63.......................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR029127(GSM416756) A549. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | GSM359187(GSM359187) HepG2_2pM_1. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM416733(GSM416733) HEK293. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189782 | SRR029126(GSM416755) 143B. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | GSM359175(GSM359175) hela_5_pct. (hela) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM359176(GSM359176) hela_chip_smrna_pure. (hela) | GSM359193(GSM359193) HepG2_3pM_7. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR038857(GSM458540) D20. (cell line) | GSM359195(GSM359195) HepG2_TAP_depl. (cell line) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | TAX577745(Rovira) total RNA. (breast) | DRR001487(DRX001041) "Hela long nuclear cell fraction, LNA(+)". (hela) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM359177(GSM359177) hela_nucl_a. (hela) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................AGCTGTGATGATCAGAAAGAAATGCATCTGAGA...................................................................................................................................................... | 33 | 1 | 220.00 | 220.00 | 100.00 | 21.00 | 13.00 | 11.00 | 11.00 | 10.00 | 5.00 | 8.00 | 7.00 | 7.00 | 6.00 | - | 3.00 | - | 4.00 | - | 1.00 | 3.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | 2.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGTGATGATCAGAAAGAAATGCATCTGAGA...................................................................................................................................................... | 30 | 1 | 11.00 | 11.00 | 7.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGCTGTGATGATCAGAAAGAAATGC.............................................................................................................................................................. | 25 | 1 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | 6.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................CAGCTGTGATGATCAGAAAGAAATGCATCTGAGA...................................................................................................................................................... | 34 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................TGATCAGAAAGAAATGCATCTGAGA...................................................................................................................................................... | 25 | 1 | 8.00 | 8.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TCAGAAAGAAATGCATCTGAGA...................................................................................................................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGCTGTGATGATCAGAAAGAAATGCATCTGATA...................................................................................................................................................... | 33 | 4.00 | 0.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................AGCTGTGATGATCAGAAAGAAATGCAT............................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ..........................................................................ATGATCAGAAAGAAATGCATCTGAGA...................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGCTGTGATGATCAGAAAGAAATGCA............................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........CCGAATTCCCTCCTCTGC............................................................................................................................................................................................................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................AGCTGTGATGATCAGAAAGAAATGCATCTGAG....................................................................................................................................................... | 32 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................ATCAGAAAGAAATGCATCTGAGA...................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................AATTTGATCAGATCG....................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CCCTGGTAGGAGATGGGGAGGCATG.................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................CTCTGTCTCTCTCTCTCTTTGTGC.................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................CAGCTGTGATGATCAGAAAGAAAT................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCCTCCCAGGGCTCCGCC............................................................................. | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................CAGCTGTGATGATCAGAAAGAAATG............................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGTGATGATCAGAAAGAAATGCA............................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGCTGTGATGATCAGAAAGAAATGCATCTGAGC...................................................................................................................................................... | 33 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGCTGTGATGATCAGAAAGAAAT................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GCTCAGAATTTGATCAGATCGACAC................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................ATAACCCTTCTGTGTGAAG................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ............................................................................................................................................................................................................................CAGATCGACACATCCAAC............ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................GAATTTGATCAGATCGACACATA................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................CAGAATTTGATCAGATCGACACATCCAACCC.......... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGCACTCTGTCTCTCTCTCTCTTTCC.................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................GAATTTGATCAGATCGACATA.................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................CAGCTGTGATGATCAGAAAGAAATGCATCTGAG....................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TCAGATCGACACATCCAACC........... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................CAGCTGTGATGATCAGAAAGAAATGC.............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAGTCCGCTCAGAATTTGATCAGATC........................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................ATCAGATCGACACATCCAACCC.......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GGAGGACATGTGAAAAGT............................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................GCACTCTGTCTCTCTCTCTC......................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AGAATTTGATCAGATCGACACA.................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TGATCAGAAAGAAATGCA............................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AGGGGGAGGACATGTG..................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ................................................................................AGAAAGAAATGCATCTG......................................................................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTCTGAGGAGCCGAATTCCCTCCTCTGTCAAATAGGAATAACCCTTCTGTGTCTACCTCCCATGGCAGCTGTGATGATCAGAAAGAAATGCATCTGAGAAAGGGGGAGGACATGTGAAAGGTGTCCCTGGTAGGAGATGGGGAGGTAGGCCATGTCCTCCCAGGGCTCCGTGGCACTCTGTCTCTCTCTCTCTTTCCAGGAGTCCGCTCAGAATTTGATCAGATCGACACATCCAACCCAAACTGTGTG .................................................................(((.((.((.....))...))...)))..(((.(((((((((((((((.(((...((...((((((.((((.((((.........)))).)))).))))))..))....)))..))))...)))))))))))))).................................................. ..............................................................63.......................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR029127(GSM416756) A549. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | GSM359187(GSM359187) HepG2_2pM_1. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM416733(GSM416733) HEK293. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189782 | SRR029126(GSM416755) 143B. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | GSM359175(GSM359175) hela_5_pct. (hela) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM359176(GSM359176) hela_chip_smrna_pure. (hela) | GSM359193(GSM359193) HepG2_3pM_7. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR038857(GSM458540) D20. (cell line) | GSM359195(GSM359195) HepG2_TAP_depl. (cell line) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | TAX577745(Rovira) total RNA. (breast) | DRR001487(DRX001041) "Hela long nuclear cell fraction, LNA(+)". (hela) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM359177(GSM359177) hela_nucl_a. (hela) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................AGTCCGCTCAGAATTTGTT.............................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................AATAACCCTTCTGTGTTATT................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |