| (1) AGO1.ip OTHER.mut | (1) AGO2.ip | (2) B-CELL | (5) BRAIN | (7) BREAST | (13) CELL-LINE | (3) HEART | (10) LIVER | (1) OTHER | (1) RRP40.ip | (16) SKIN | (2) UTERUS |
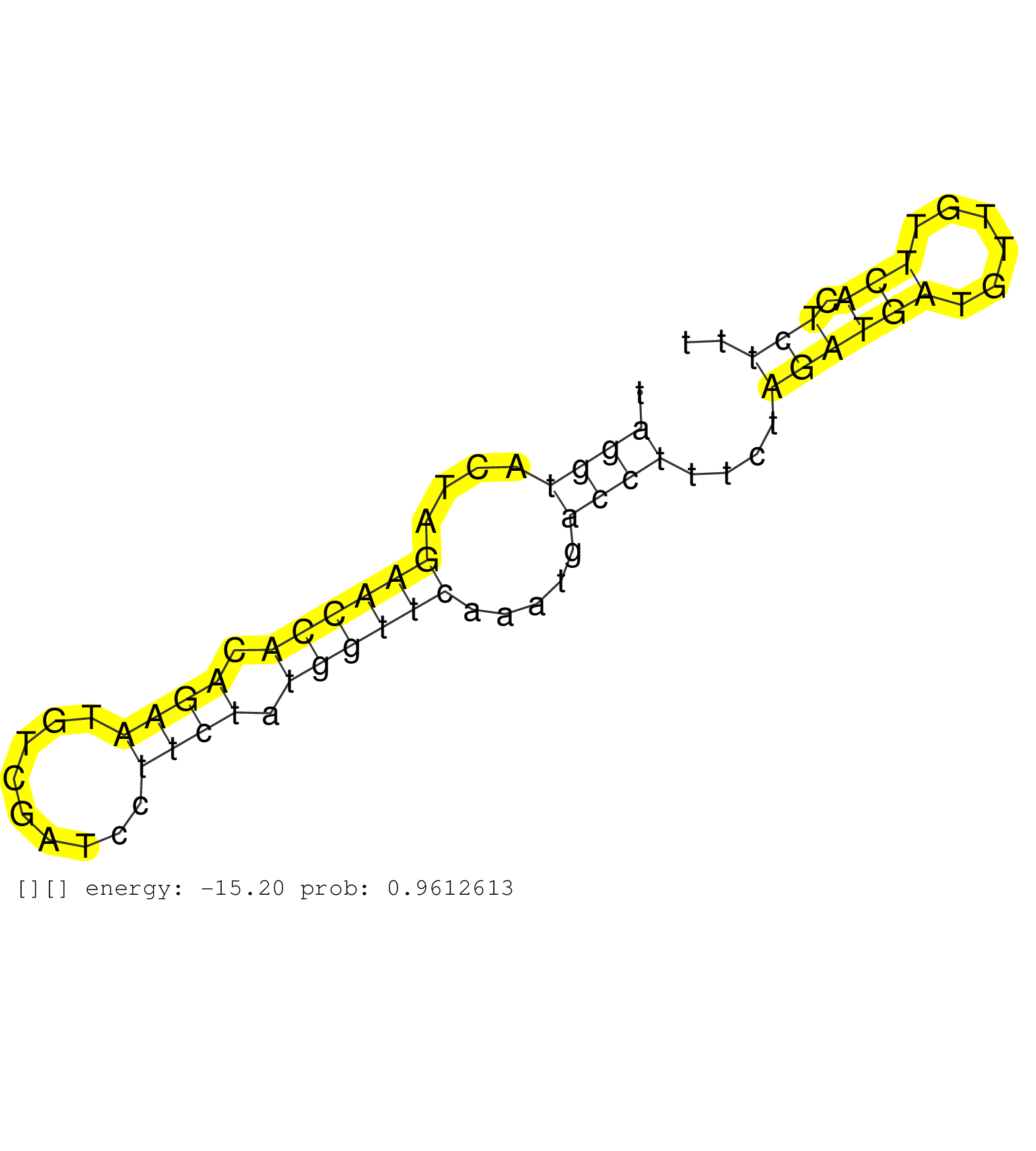
| TCTTATAAATTAGGGATCCCAAGCTGATTTCTAAACATTTCTACTGAGTAAAGAAATTATACCAAATATTTGATTAGCTCATTCTATTTAATTTTTGTTTTTGTTTTGTATCATGGATTAGGTACTAGAACCACAGAATGTCGATCCTTCTATGGTTCAAATGACCTTTCTAGATGATGTTGTTCACTCTTTGTTAAAAGGTGAAAATATTGGCATTACATCACGACGCAGGTCTCGTGCCAATCAAAAC .......................................................................................................................((((....((((((.((((.........)))).)))))).....))))....((((((......))).)))............................................................ ......................................................................................................................119......................................................................192........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | TAX577739(Rovira) total RNA. (breast) | TAX577740(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR343334 | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................ACTAGAACCACAGAATGTCGATC........................................................................................................ | 23 | 1 | 16.00 | 16.00 | 1.00 | - | - | - | - | - | - | - | 3.00 | 1.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCGAT......................................................................................................... | 22 | 1 | 12.00 | 12.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCGATCC....................................................................................................... | 24 | 1 | 9.00 | 9.00 | 1.00 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCG........................................................................................................... | 20 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCGATCCT...................................................................................................... | 25 | 1 | 7.00 | 7.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCGA.......................................................................................................... | 21 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGATGATGTTGTTCACT.............................................................. | 17 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CTCATTCTATTTAATTTTTGTT....................................................................................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCGATA........................................................................................................ | 23 | 1 | 3.00 | 12.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................AGATGATGTTGTTCACTCTTTG......................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGATGATGTTGTTCACTCT............................................................ | 19 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGATGATGTTGTTCACTC............................................................. | 18 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................AAAATATTGGCATTACATCACG......................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................ATTACATCACGACGCAGGTA................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................AATGACCTTTCTAGATGATG....................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGTTAAAAGGTGAAAATAT........................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TAGAACCACAGAATGTCGATCC....................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGGATTAGGTACTAGAACCACAGA................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TACTAGAACCACAGAATGTCGA.......................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................CTTTCTAGATGATGTTGTTCACT.............................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTGTTTTTGTTTTGTATCATGGATTAG................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CTAGAACCACAGAATGT............................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GTACTAGAACCACAGAATGTCGATCC....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATGTCGATCCTTCTATGGT.............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGATGATGTTGTTCACTCTT........................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................GAACCACAGAATGTCGATCC....................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................AGGTACTAGAACCACAGAATGT............................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CTCATTCTATTTAATTTTTGTTTTTGTA................................................................................................................................................. | 28 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................AATTTTTGTTTTTGTTTCTTG............................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................TAGAACCACAGAATGTCGATCCT...................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AACCACAGAATGTCGATCT....................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................TTTCTAGATGATGTTGTTCACT.............................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CTAGAACCACAGAATGTCGA.......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCGATCCTT..................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACTAGAACCACAGAATGTCGATCT....................................................................................................... | 24 | 1 | 1.00 | 16.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TACTAGAACCACAGAATG.............................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................CTCATTCTATTTAATTCTC.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| ........................................................................................................................GGTACTAGAACCACAGAATGTC............................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CTATTTAATTTTTGTAGAG.................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................GGTACTAGAACCACAGAATGTCG........................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................ATTTTTGTTTTTGTTTTGTAGCCG........................................................................................................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................TACTAGAACCACAGAATGTCGATCC....................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GTTCAAATGACCTTTCTAGATGATGTTGT................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CCTTCTATGGTTCAAATGACCTTTC................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................AGGTACTAGAACCACAGAATGTCGAAAA....................................................................................................... | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....ATAAATTAGGGATCCAAA.................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................TTAGGTACTAGAACCACAGAATGTC............................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGATGATGTTGTTCACTCTTT.......................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TTCTATTTAATTTTTGTTTTTGTTTTGCATC.......................................................................................................................................... | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTTATAAATTAGGGATCCCAAGCTGATTTCTAAACATTTCTACTGAGTAAAGAAATTATACCAAATATTTGATTAGCTCATTCTATTTAATTTTTGTTTTTGTTTTGTATCATGGATTAGGTACTAGAACCACAGAATGTCGATCCTTCTATGGTTCAAATGACCTTTCTAGATGATGTTGTTCACTCTTTGTTAAAAGGTGAAAATATTGGCATTACATCACGACGCAGGTCTCGTGCCAATCAAAAC .......................................................................................................................((((....((((((.((((.........)))).)))))).....))))....((((((......))).)))............................................................ ......................................................................................................................119......................................................................192........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | TAX577739(Rovira) total RNA. (breast) | TAX577740(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR343334 | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................GAGTAAAGAAATTATTCTA.......................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |