| (12) B-CELL | (1) BRAIN | (3) BREAST | (3) CELL-LINE | (2) CERVIX | (1) HEART | (3) LIVER | (2) OTHER | (4) SKIN | (1) UTERUS |
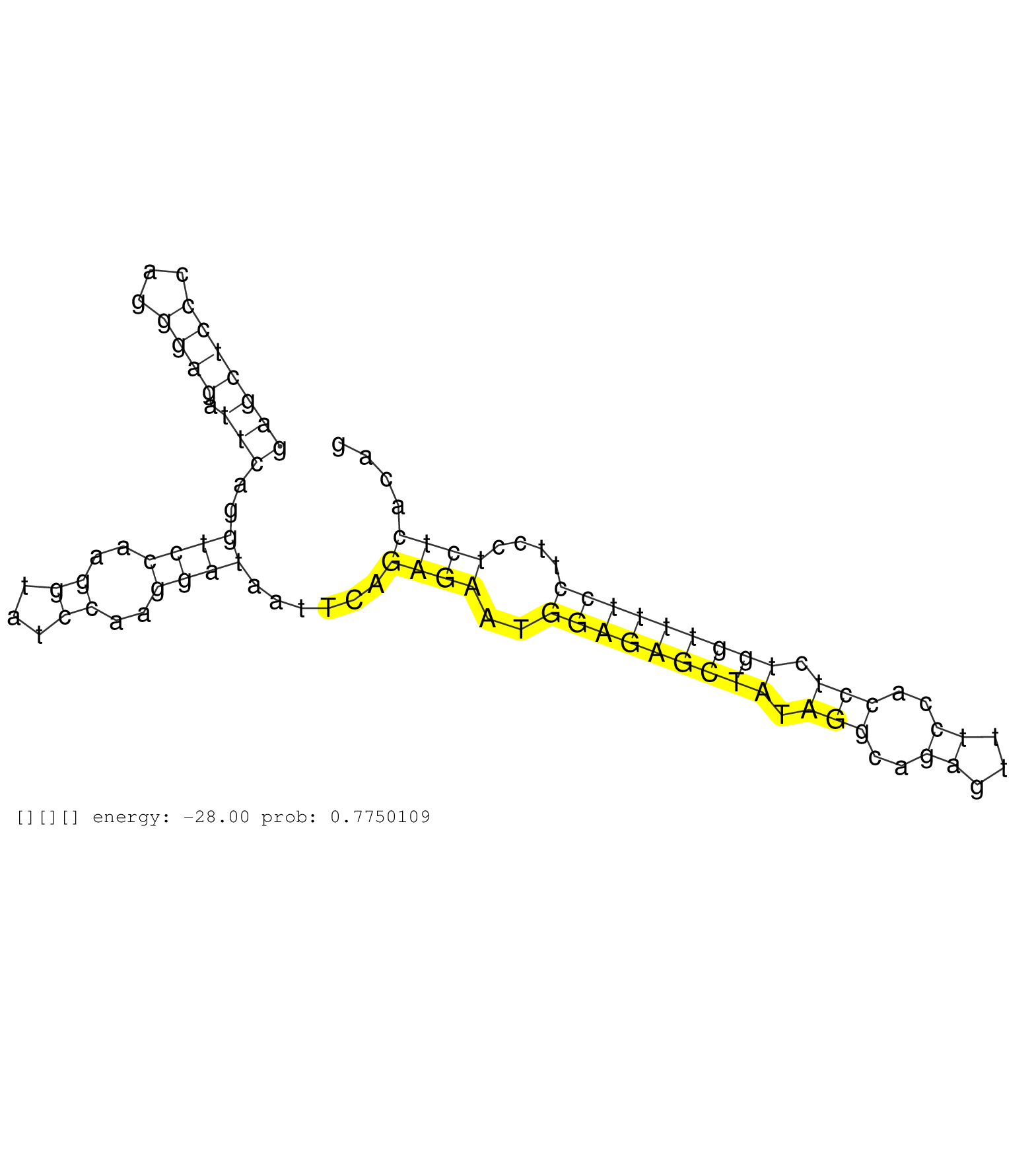
| TATTATTATTATATAGCTATGGTAAAGCAGACCTAGGTAGTTCTCTTTTCCAGATACGAATGAGGATTCTGCTCTGAATTACTGCTTTCTCTCCAGTCTGGAGCTCCCAGGGAGATTCAGGTCCAAGGTATCCAAGGATAATTCAGAGAATGGAGAGCTATAGGCAGAGTTTCCACCTCTGGTTTTCCTTCCTCTCACAGAGCACCTGTCAAAGCCTAAAGTCACCATGGGTCTGCAGAGCAATAAGAAT .................................................................................................................................................((((..(((((((((.(((..((...))..))).)))))))))....))))...................................................... .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR189782 | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR038859(GSM458542) MM386. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR040010(GSM532895) G529N. (cervix) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040029(GSM532914) G026T. (cervix) | SRR189787 | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | TAX577740(Rovira) total RNA. (breast) | SRR189785 | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................TCAGAGAATGGAGAGCTATAG....................................................................................... | 21 | 1 | 14.00 | 14.00 | 8.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCAGAGAATGGAGAGCTATAGG...................................................................................... | 22 | 1 | 10.00 | 10.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................TTCAGAGAATGGAGAGCTA.......................................................................................... | 19 | 1 | 8.00 | 8.00 | - | - | - | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGAGAATGGAGAGCTATAGGCA.................................................................................... | 22 | 1 | 6.00 | 6.00 | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................CAAGGATAATTCAGAGAATGGAG............................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTCAGAGAATGGAGAGCTATAG....................................................................................... | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CTAAAGTCACCATGGGTCTGCAGA........... | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGAGAATGGAGAGCTATAGGCAT................................................................................... | 23 | 1 | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................TCAGAGAATGGAGAGCTATA........................................................................................ | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAGAGAATGGAGAGCTATAGG...................................................................................... | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................AAAGTCACCATGGGTCTGCAG............ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGAGAATGGAGAGCTATAGGCAA................................................................................... | 23 | 1 | 1.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GAGAGCTATAGGCAGACGA............................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................CAGAGAATGGAGAGCTATAGGAT.................................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................CCATGGGTCTGCAGAGTT........ | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................AAAGTCACCATGGGTCTGCAGAG.......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TCCAAGGATAATTCAGAGAATGG................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTCAGAGAATGGAGAGCTATAA....................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................CACCATGGGTCTGCAGAGCAATAAG... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCAGAGAATGGAGAGCTATAGGT..................................................................................... | 23 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAGAGAATGGAGAGCTAT......................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCAGAGAATGGAGAGCTATAGGA..................................................................................... | 23 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTCTGGTTTTCCTTCGTGT....................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TTCAGAGAATGGAGAGCTATAGGTT.................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGAGAATGGAGAGCTATAGG...................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TCACAGAGCACCTGTCTCT..................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TTCAGAGAATGGAGAGCTATAGG...................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................TCAGAGAATGGAGAGCTATAA....................................................................................... | 21 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGAGAATGGAGAGCTATA........................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TCCAAGGATAATTCAGAGAATGGA................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAGAGAATGGAGAGCTATA........................................................................................ | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TATTATTATTATATAGCTATGGTAAAGCAGACCTAGGTAGTTCTCTTTTCCAGATACGAATGAGGATTCTGCTCTGAATTACTGCTTTCTCTCCAGTCTGGAGCTCCCAGGGAGATTCAGGTCCAAGGTATCCAAGGATAATTCAGAGAATGGAGAGCTATAGGCAGAGTTTCCACCTCTGGTTTTCCTTCCTCTCACAGAGCACCTGTCAAAGCCTAAAGTCACCATGGGTCTGCAGAGCAATAAGAAT .................................................................................................................................................((((..(((((((((.(((..((...))..))).)))))))))....))))...................................................... .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR189782 | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR038859(GSM458542) MM386. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR040010(GSM532895) G529N. (cervix) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040029(GSM532914) G026T. (cervix) | SRR189787 | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | TAX577740(Rovira) total RNA. (breast) | SRR189785 | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................ATACGAATGAGGATTAGT................................................................................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................ATACGAATGAGGATTAAA................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ...............................................................................TACTGCTTTCTCTCCACCTG....................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................CCATGGGTCTGCAGAAAC........ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................TTCCTTCCTCTCACAGGTA............................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................TACTGCTTTCTCTCCACTTG....................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................GAGCTCCCAGGGAGATGACT.................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ................................................................................ACTGCTTTCTCTCCACCCT....................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................TACTGCTTTCTCTCCACCTC....................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................CCTTGGACCTGAATCTCCC.......................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGGTGGAAACTCTGC........................................................................ | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |