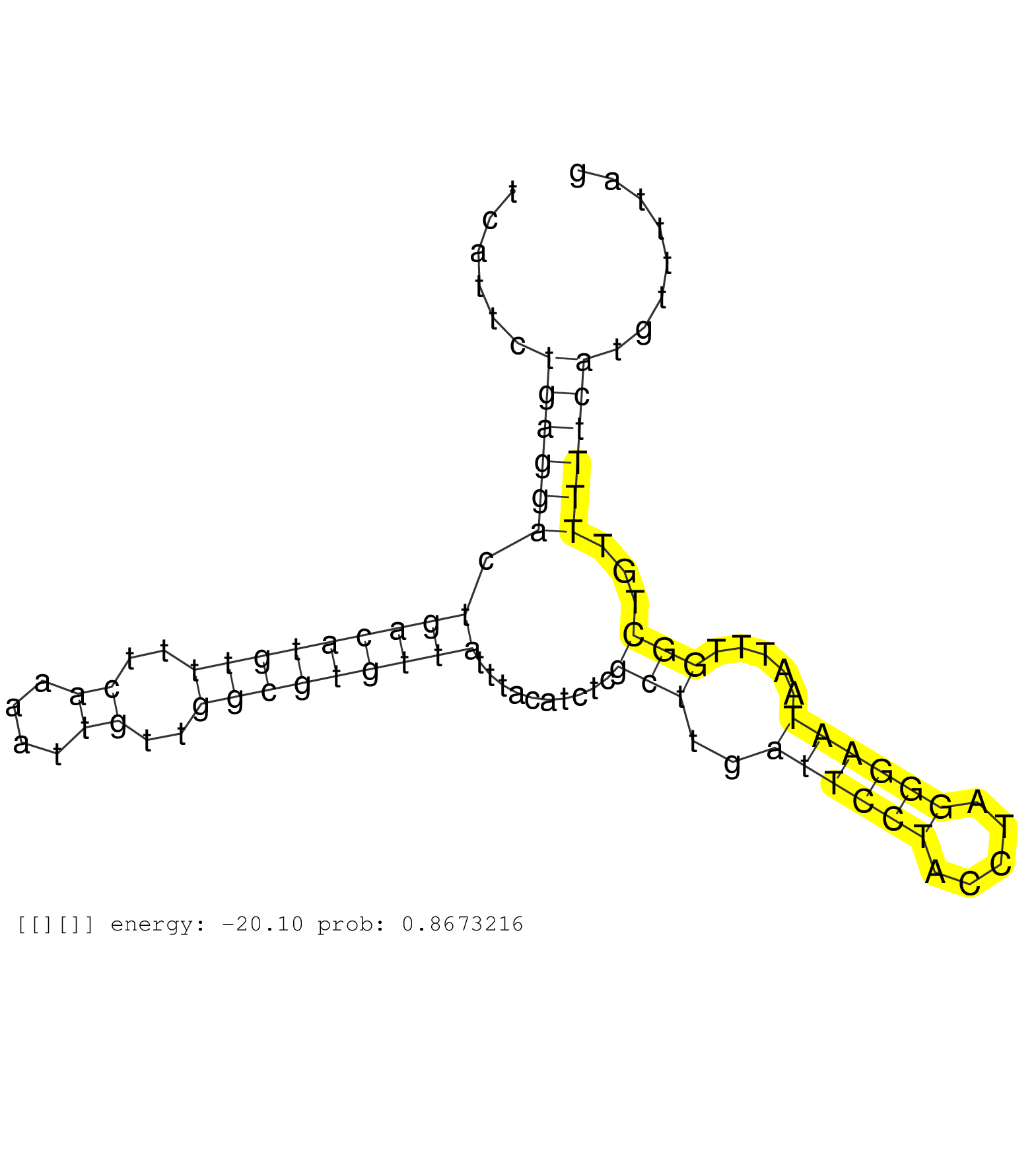
| (1) BREAST | (11) CELL-LINE | (2) HELA | (1) LIVER | (3) OTHER | (3) SKIN | (1) TESTES | (1) XRN.ip |
| GGCCGTGGTCGTGGTGGGCGCTTTGGTTCCAGAGGAGGCCCAGGAGGAGGGTGAGTACCAGCTTTTAGCATCTCTGCACTTGGTTGTTAGTTTCTGCTTGTCATCTAAATAGGAGTTTTAGAGTAGACTAATGTCACTTGGTTTTGCGAGAGCCAACTGTGGTCTTTTCCTGCACTCCTGTTAAATGAATTTGGCCCCACTATGCTAAGAATGATTCTTTCCCTCCCCGGAGACTTTGTCACTCATTCTGAGGACTGACATGTTTTCAAAATTGTTGGCGTGTTATTTACATCTCGCTTGATTCCTACCTAGGGAATAATTTGGCTGTTTTTCATGTTTTAGGTTCAGGCCCTTTGTACCACATATCCCATTTGACTTCTATTTGGTGAGTA ...................................................................................................................................................((.((((((.(((((......)).))).((((.......(((((.(((.........((.(((((((.....(((......)))(((...)))..))))))).))...(((((((((..((....))..)))))))))..........))).))))).......)))).....)))))).))............................................................... ...................................................................................................................................................148.......................................................................................................................................................................................334........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037939(GSM510477) 293cand5_rep1. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189786 | SRR343334 | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR189782 | TAX577742(Rovira) total RNA. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................................................................TCCTACCTAGGGAATAATTTGGCTGTTTT............................................................. | 29 | 1 | 20.00 | 20.00 | 15.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................ATTGTTGGCGTGTTATTTACATCTCGCTTGATTCCTACCTAGGGAATAATTTGGCTGTTTTTCATGTTTTA................................................... | 71 | 1 | 10.00 | 10.00 | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................TCCTACCTAGGGAATAATTTGGCTGGTTT............................................................. | 29 | 6.00 | 0.00 | 1.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................AATTGTTGGCGTGTTATTTACATCTCGCTTGATTCCTACCTAGGGAATAATTTGGCTGTTTTTCATGTTTTAG.................................................. | 73 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................ATTGTTGGCGTGTTATTTACATCTCGCTTGATTCCTACCTAGGGAATAATTTGGCTGTTTTTCATGTTTTAG.................................................. | 72 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GCATCTCTGCACTTGGTTGTTAGTTTCTTC....................................................................................................................................................................................................................................................................................................... | 30 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | |
| .....................................................................................................................................................................................................................................................................................................................TAGGGAATAATTTGGCTGTTT.............................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................TGGTTCCAGAGGAGGCC................................................................................................................................................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............TGGGCGCTTTGGTTCCAGAGGAGGCC................................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .GCCGTGGTCGTGGTGGGCGCTTTGGTTCCAGAGG..................................................................................................................................................................................................................................................................................................................................................................... | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................GGGAATAATTTGGCTGTTTTT............................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................CCACATATCCCATTTGACTTCTACTT........ | 26 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................GGTTTTGCGAGAGCCAACTGTGGTCTTTTCC.............................................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................TTCAGGCCCTTTGTACCACATCCC......................... | 24 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................AATTTGGCTGTTTTTCATGTTTTAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TTCCAGAGGAGGCCCAGGAGGAGGGCT................................................................................................................................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................................................................................................GGAATAATTTGGCTGTTTTTCATGTTTTAG.................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GTGGTCGTGGTGGGCGCCCG................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................TCCTACCTAGGGAATAATTTGGCTGATTT............................................................. | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................CCAACTGTGGTCTTTTTTT............................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................................................................................................................................................ACCACATATCCCATTTGTTTG.............. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| .....................................................................................................................................................................................................................................................................GTTTTCAAAATTGTTGGCGTGTTATTTAC...................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................TTAGAGTAGACTAATGT.................................................................................................................................................................................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| GGCCGTGGTCGTGGTGGGCGCTTTGGTTCCAGAGGAGGCCCAGGAGGAGGGTGAGTACCAGCTTTTAGCATCTCTGCACTTGGTTGTTAGTTTCTGCTTGTCATCTAAATAGGAGTTTTAGAGTAGACTAATGTCACTTGGTTTTGCGAGAGCCAACTGTGGTCTTTTCCTGCACTCCTGTTAAATGAATTTGGCCCCACTATGCTAAGAATGATTCTTTCCCTCCCCGGAGACTTTGTCACTCATTCTGAGGACTGACATGTTTTCAAAATTGTTGGCGTGTTATTTACATCTCGCTTGATTCCTACCTAGGGAATAATTTGGCTGTTTTTCATGTTTTAGGTTCAGGCCCTTTGTACCACATATCCCATTTGACTTCTATTTGGTGAGTA ...................................................................................................................................................((.((((((.(((((......)).))).((((.......(((((.(((.........((.(((((((.....(((......)))(((...)))..))))))).))...(((((((((..((....))..)))))))))..........))).))))).......)))).....)))))).))............................................................... ...................................................................................................................................................148.......................................................................................................................................................................................334........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037939(GSM510477) 293cand5_rep1. (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | DRR001485(DRX001039) "Hela long total cell fraction, LNA(+)". (hela) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189786 | SRR343334 | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR189782 | TAX577742(Rovira) total RNA. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................CAGAGGAGGCCCAGGATAGT....................................................................................................................................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................................................................................................................TACCACATATCCCATTTA.................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |