| (5) B-CELL | (3) BRAIN | (5) BREAST | (9) CELL-LINE | (12) CERVIX | (1) FIBROBLAST | (7) HEART | (2) HELA | (1) KIDNEY | (6) LIVER | (1) OTHER | (22) SKIN | (1) TESTES | (1) XRN.ip |
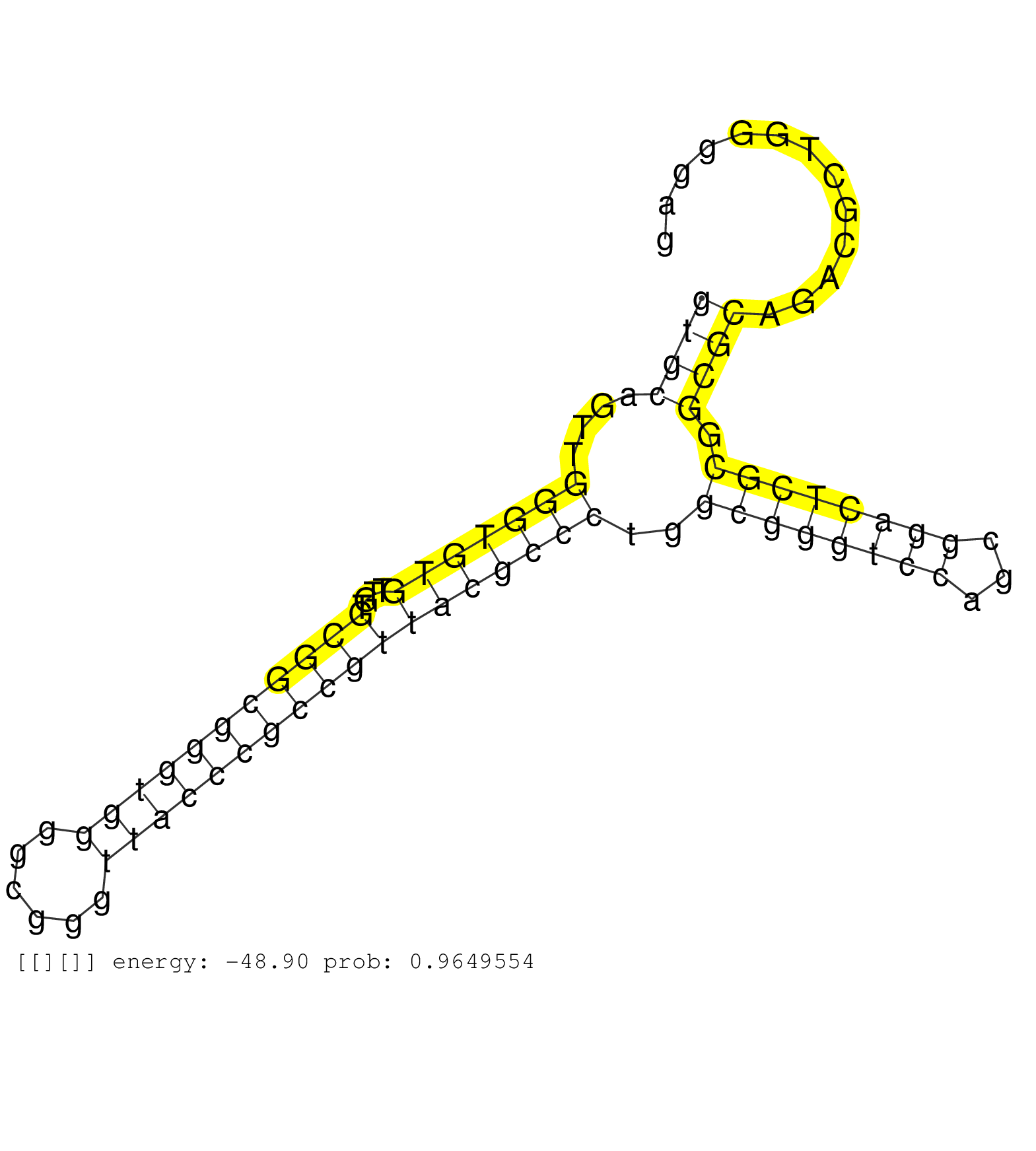
| TATACCCTGGTACTTCTAGTCTAAAACAGGATTTGACTCTACTAATCCAGGTTAGTCTTGCCTTTCCCATTACTCATGTGCTTTCGTTGAAACCAAAGACTTTTTCTTTTGTGCCCATTTCTACTCACTGCCTGGCCGTCTGCACGACGATCTGTGTTTCCTGGTAGTTCTCCAGTCTAGTAGCTACTATAAACTACTCTCCCTATTTCCCGCTCTGTGGAGATGTCGTTCCAACAGGTCCTCTCCACTC ................................................................................................(((((.........(((........)))..((((((.((......(((((......)))))..)).)))))).....))))).((((..........))))..................................................... .........................................................................................90............................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM532880(GSM532880) G659T. (cervix) | GSM532876(GSM532876) G547T. (cervix) | GSM532874(GSM532874) G699T. (cervix) | GSE20417(GSM514985) Colon and lung cancer pool. | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | TAX577580(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040019(GSM532904) G701T. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR040037(GSM532922) G243T. (cervix) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR191400(GSM715510) 37genomic small RNA (size selected RNA from t. (breast) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR040031(GSM532916) G013T. (cervix) | SRR040007(GSM532892) G601T. (cervix) | GSM532890(GSM532890) G576T. (cervix) | SRR040039(GSM532924) G531T. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR040025(GSM532910) G613T. (cervix) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM532886(GSM532886) G850T. (cervix) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................AAAGACTTTTTCTTTGACC......................................................................................................................................... | 19 | 47.00 | 0.00 | 5.00 | 4.00 | 5.00 | 6.00 | 6.00 | 4.00 | - | 2.00 | - | - | 2.00 | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AAAGACTTTTTCTTTTATC......................................................................................................................................... | 19 | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AAAGACTTTTTCTTTTACCA........................................................................................................................................ | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AAAGACTTTTTCTTTTACC......................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................AAGACTTTTTCTTTTACCA........................................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AAAGACTTTTTCTTTGAAC......................................................................................................................................... | 19 | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................ACAGGATTTGACTCTACTAAT............................................................................................................................................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AAAGACTTTTTCTTTGATC......................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AAAGACTTTTTCTTTTACCT........................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................AACAGGATTTGACTCTACTAATC........................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................GCTCTGTGGAGATGTCGTTC................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AAAGACTTTTTCTTTGACG......................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......TGGTACTTCTAGTCTAAAACCC............................................................................................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................ATGTCGTTCCAACAGGTC.......... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TCTAAAACAGGATTTGACTCTACT............................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................GTCTAGTAGCTACTATAAAC........................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTGCCCATTTCTACTCACTGCCTGGC.................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CCAAAGACTTTTTCTCTGA........................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................CTAAAACAGGATTTGACTCTACTAA............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCCCGCTCTGTGGAGATG......................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AAAGACTTTTTCTTTGAGT......................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................AACAGGATTTGACTCTACTA.............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AAAGACTTTTTCTTTTAC.......................................................................................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................TAAAACAGGATTTGACTCTACTAA............................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAACCAAAGACTTTTGGT............................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........ACTTCTAGTCTAAAATG.............................................................................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................GCTCTGTGGAGATGTCGTT.................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................CCCATTTCTACTCACTTG....................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................CTAAAACAGGATTTGATGGG.................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AAAGACTTTTTCTTTTAAT......................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................AAAGACTTTTTCTTTGAGC......................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................CCAAAGACTTTTTCTTTTG........................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TATACCCTGGTACTTCTAGTCTAAAACAGGATTTGACTCTACTAATCCAGGTTAGTCTTGCCTTTCCCATTACTCATGTGCTTTCGTTGAAACCAAAGACTTTTTCTTTTGTGCCCATTTCTACTCACTGCCTGGCCGTCTGCACGACGATCTGTGTTTCCTGGTAGTTCTCCAGTCTAGTAGCTACTATAAACTACTCTCCCTATTTCCCGCTCTGTGGAGATGTCGTTCCAACAGGTCCTCTCCACTC ................................................................................................(((((.........(((........)))..((((((.((......(((((......)))))..)).)))))).....))))).((((..........))))..................................................... .........................................................................................90............................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM532880(GSM532880) G659T. (cervix) | GSM532876(GSM532876) G547T. (cervix) | GSM532874(GSM532874) G699T. (cervix) | GSE20417(GSM514985) Colon and lung cancer pool. | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | TAX577580(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040019(GSM532904) G701T. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR040037(GSM532922) G243T. (cervix) | SRR191586(GSM715696) 197genomic small RNA (size selected RNA from . (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR191400(GSM715510) 37genomic small RNA (size selected RNA from t. (breast) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR040031(GSM532916) G013T. (cervix) | SRR040007(GSM532892) G601T. (cervix) | GSM532890(GSM532890) G576T. (cervix) | SRR040039(GSM532924) G531T. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR040025(GSM532910) G613T. (cervix) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM532886(GSM532886) G850T. (cervix) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................GAAACCAAAGACTTTCGG................................................................................................................................................ | 18 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....AGACTAGAAGTACCAGGG.................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GACGATCTGTGTTTCCC........................................................................................ | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................AGGCAGTGAGTAGAAATGGGCA..................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TCCAACAGGTCCTCTCCC... | 18 | 1.29 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.29 | - | - | - | 0.14 | 0.29 | 0.29 | - | - | - | 0.14 | - | - | - | - | 0.14 | - | - | |
| ...........................................................................................................................................ACACAGATCGTCGTGCAG............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TTTAGACTAGAAGTACCA................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCAGTGAGTAGAAATGGGCACA....................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................AGTTCTCCAGTCTAGGCG................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................AGTCTTGCCTTTCCCGGTA.................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGGAAACACAGATCGTCGTGCA........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACAGATCGTCGTGCAG............................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GAAACCAAAGACTTTCGGG............................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................CTACTCTCCCTATTTCGAA...................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................GACGATCTGTGTTTCCG........................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................ACTATAAACTACTCTCCCTG............................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................AAACACAGATCGTCGTG........................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................AGAGGACCTGTTGGA...... | 15 | 7 | 0.86 | 0.86 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.43 | - | - | - | - | - | - | 0.14 | - | 0.14 | - | - | - | - | 0.14 | - |
| .....................................................................................................................................................................................................................................TCCAACAGGTCCTCTCC.... | 17 | 0.86 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.29 | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | |
| .....................................................................................................................................................................................................................................TCCAACAGGTCCTCTC..... | 16 | 0.57 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | 0.14 | - | - | - | 0.14 | 0.14 | - | - | - | - | |
| ...............................................................................................................................................................................................AAATAGGGAGAGTAGTT.......................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CGGGAAATAGGGAGA...................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TCCAACAGGTCCTCTTCA... | 18 | 0.14 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |