| (3) B-CELL | (1) BRAIN | (2) BREAST | (18) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (2) HELA | (2) LIVER | (1) OTHER | (7) SKIN | (1) XRN.ip |
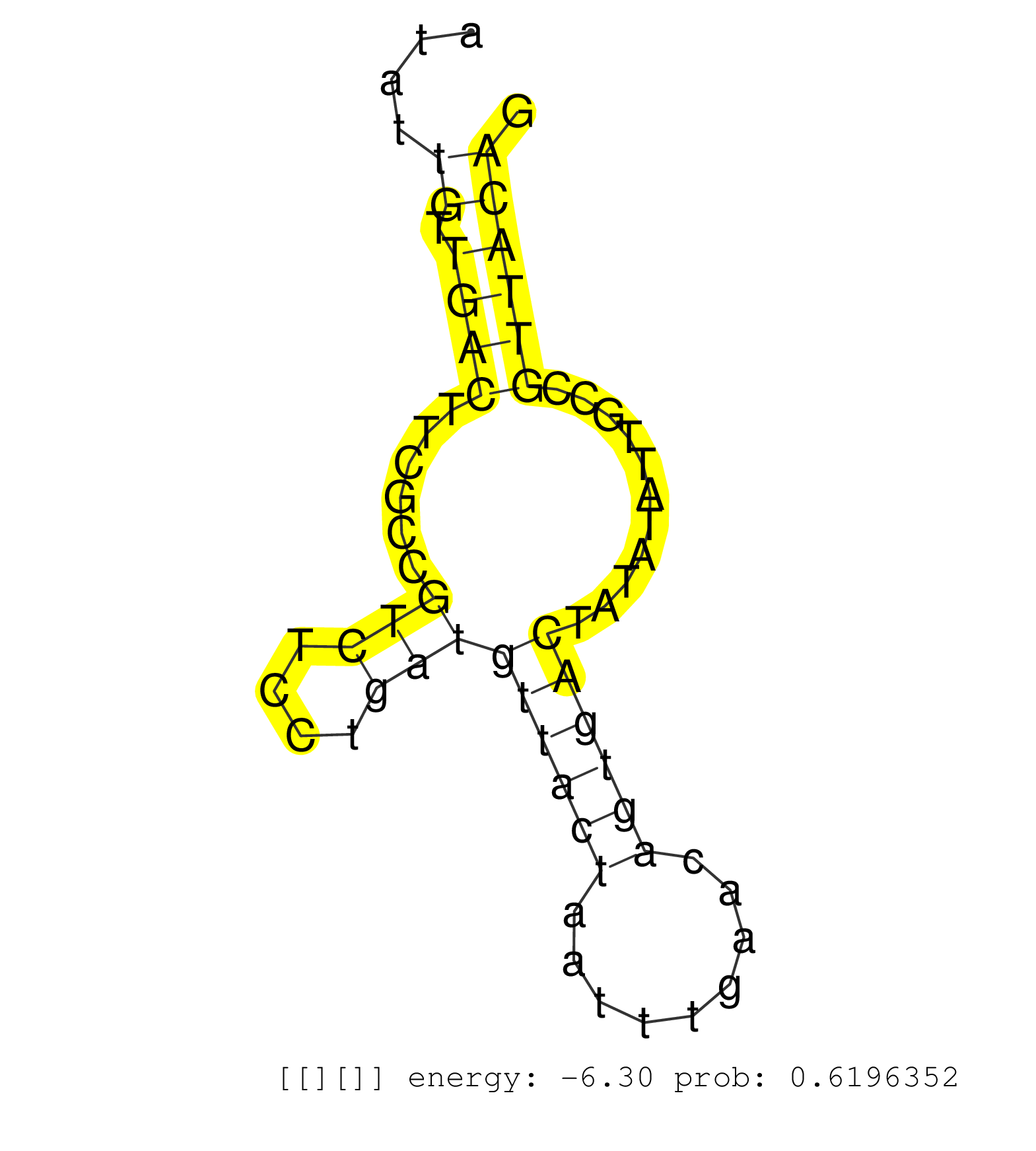
| GATCGCACCACTCCATTCTAGCCTGGGCGAAAGAGTGAGACTCTATTTATAAAAAAAGAAAGAAAGAAAAGACCCTTAATGACTTTTTCCCCTCTCTTAAAACCTGGTTGTATTAGTGACTTTGACATACATGTATATTGTTGACTTCGCCGTCTCCTGATGTTACTAATTTGAACAGTGACTATATATTGCCGTTACAGACCAGAAGGGAAAGATTTCAGCATTCATCGACTTGTCCTGGGGACACACA .........................................................................................................(((.((((((((.(((....((.....((((((((..(((......)))...)))))).)).....))...))).)))).)))).)))......................................................... .............................................................................78........................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR029127(GSM416756) A549. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR040031(GSM532916) G013T. (cervix) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR040035(GSM532920) G001T. (cervix) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM359196(GSM359196) m3m7G_ip. (cell line) | SRR191594(GSM715704) 70genomic small RNA (size selected RNA from t. (breast) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM956925PAZD5 | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................ACTATATATTGCCGTTACAG.................................................. | 20 | 1 | 20.00 | 20.00 | - | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................GACATACATGTATATTGTTGACTTCGCCGTCTCCTGATGTTACTAATTTGAACAGTGACTATATATTGCCGTT...................................................... | 73 | 1 | 14.00 | 14.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TTGAACAGTGACTATATATTGCCGTTACAG.................................................. | 30 | 1 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GCATTCATCGACTTGTCCTGGGGACACT.. | 28 | 4.00 | 0.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................ATTTGAACAGTGACTATATATTGCCGTTACAG.................................................. | 32 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GTGACTATATATTGCCGTTACAG.................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TTGAACAGTGACTATATATTGCCGTTACAGA................................................. | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GAAGGGAAAGATTTCAGCATTCATA..................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GACTATATATTGCCGTTACAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TTGAACAGTGACTATATATTGCCGTTACA................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGAACAGTGACTATATATTGCCGTTACAG.................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ACTATATATTGCCGTTACAT.................................................. | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................CTTTTTCCCCTCTCTAAAA..................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................TCTAGCCTGGGCGAAAGAGTGAGACCCCA............................................................................................................................................................................................................. | 29 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................GTTGACTTCGCCGTCTCC............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGAACAGTGACTATATATTGCCGTTAC.................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CTCCATTCTAGCCTGGGCCACA.......................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................CGACTTGTCCTGGGGAC..... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ACCAGAAGGGAAAGATTTCAGCATTCATCGA................... | 31 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AGGGAAAGATTTCAGCATT......................... | 19 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - |
| .......................................................................................................................................................................................................................................CTTGTCCTGGGGACACA.. | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - |
| .......................................................................................................................................................................................................GACCAGAAGGGAAAGATTTCAGCA........................... | 24 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - |
| .............................................................................................................................................................................................................AAGGGAAAGATTTCAGCA........................... | 18 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - |
| ..........................................................................................................................................................................................................CAGAAGGGAAAGATTTCAGC............................ | 20 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| .....................................................................................................................................................................................................CAGACCAGAAGGGAAAGATTTCAG............................. | 24 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| GATCGCACCACTCCATTCTAGCCTGGGCGAAAGAGTGAGACTCTATTTATAAAAAAAGAAAGAAAGAAAAGACCCTTAATGACTTTTTCCCCTCTCTTAAAACCTGGTTGTATTAGTGACTTTGACATACATGTATATTGTTGACTTCGCCGTCTCCTGATGTTACTAATTTGAACAGTGACTATATATTGCCGTTACAGACCAGAAGGGAAAGATTTCAGCATTCATCGACTTGTCCTGGGGACACACA .........................................................................................................(((.((((((((.(((....((.....((((((((..(((......)))...)))))).)).....))...))).)))).)))).)))......................................................... .............................................................................78........................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) | SRR029127(GSM416756) A549. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR040031(GSM532916) G013T. (cervix) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR040035(GSM532920) G001T. (cervix) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM359196(GSM359196) m3m7G_ip. (cell line) | SRR191594(GSM715704) 70genomic small RNA (size selected RNA from t. (breast) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM956925PAZD5 | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................CCTTAATGACTTTTTCCCAA............................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................ATTTATAAAAAAAGAATTAG.......................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................TATTTATAAAAAAAGCACT............................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................AAAAAAGAAAGAAAGAAAAGAAAAG.............................................................................................................................................................................. | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .................................................TAAAAAAAGAAAGAAAGCCCC.................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................GGGAAAGATTTCAGCATACGG...................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................AACAGTGACTATATATGTAT......................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................AAAAAAGAAAGAAAGAAAAGCGG................................................................................................................................................................................ | 23 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................GAAAGAAAGAAAAGACCAAT............................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................ATAAAAAAAGAAAGAAAGATCG.................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................AAAGAAAGAAAGAAAAGAAAAG.............................................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................AAAAAAAGAAAGAAAGAAAAGAGGAC.............................................................................................................................................................................. | 26 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................TAAAAAAAGAAAGAAAGACGAG................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................TCTTTCTTTTTTTATAAATA........................................................................................................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................AATCTTTCCCTTCTGGTCTG................................. | 20 | 7 | 0.29 | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.29 | - | - | - | - | - | - |