| (3) BREAST | (2) CELL-LINE | (12) CERVIX | (1) LIVER | (2) OTHER | (2) SKIN |
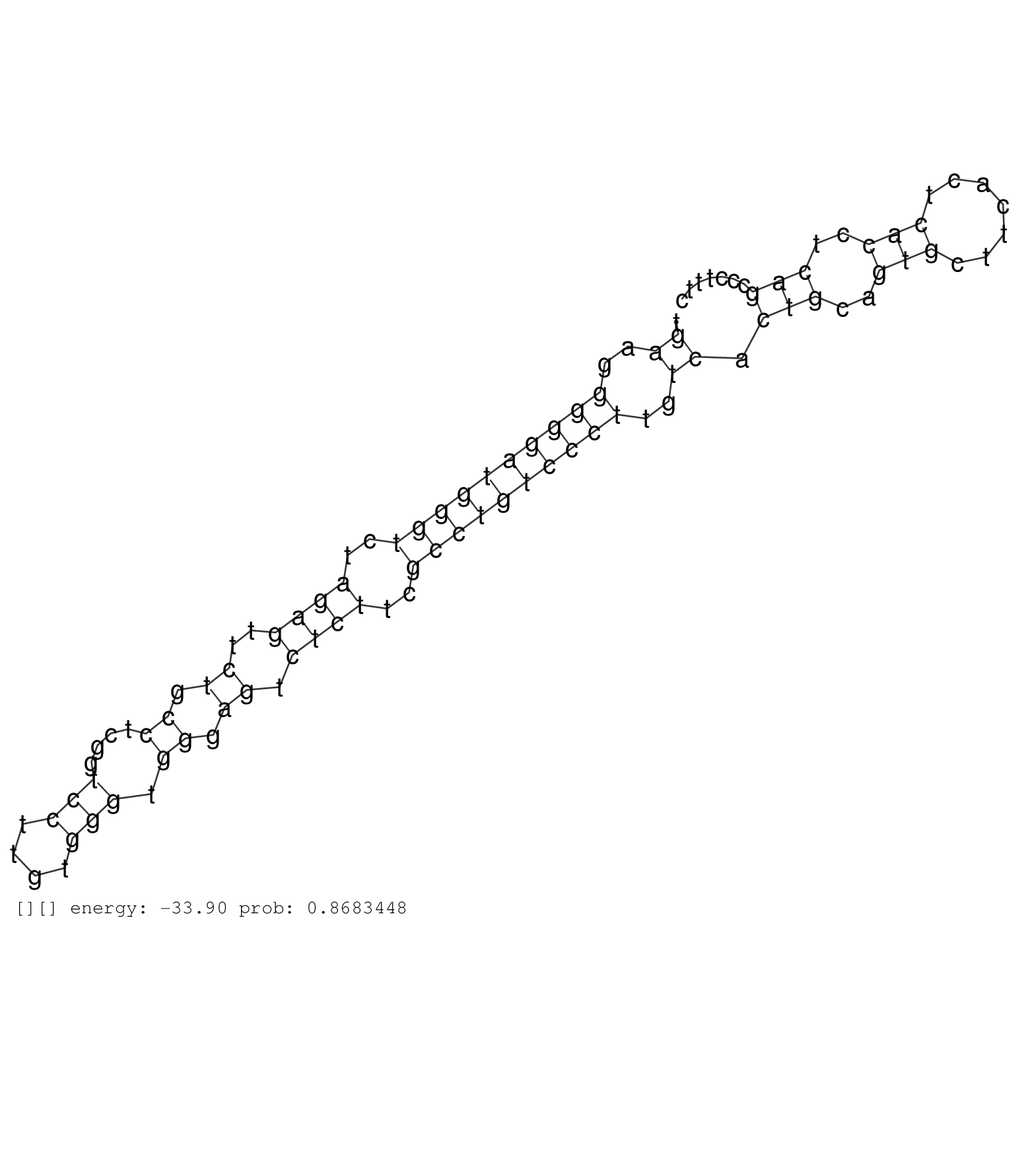
| TTTTAACTGCCTCTGGAAGTCTATAAACACATGTATAAACAAAAAAGTGAGATATTTTCTACCACATAGTCTTTCTATCTGGTGTCCAAGCTGTATTCTGTTAATTAGTTTGAAAGACTATACAGTGAGATGTGCAGTTTTCCCATTCCATTTTCGAAATGTTCTAACTTTTTAACCTGGTGATATTATCTTTCTTTCAGTTATGACATGCACCGATTACATCCCTCGCTCATCCAATGATTATACCTCA ..............................................................................................................................((((.(((.(((......))).))))))).....................((((.((.........))..)))).................................................. ..............................................................................................................................127......................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR040029(GSM532914) G026T. (cervix) | SRR040043(GSM532928) G428T. (cervix) | SRR040015(GSM532900) G623T. (cervix) | SRR040011(GSM532896) G529T. (cervix) | GSM532876(GSM532876) G547T. (cervix) | SRR040031(GSM532916) G013T. (cervix) | SRR037937(GSM510475) 293cand2. (cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR040025(GSM532910) G613T. (cervix) | SRR189782 | SRR189785 | SRR040039(GSM532924) G531T. (cervix) | SRR040037(GSM532922) G243T. (cervix) | SRR040016(GSM532901) G645N. (cervix) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040022(GSM532907) G575N. (cervix) | TAX577590(Rovira) total RNA. (breast) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR191445(GSM715555) 110genomic small RNA (size selected RNA from . (breast) | TAX577745(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................GTGCAGTTTTCCCATGAAT.................................................................................................... | 19 | 14.00 | 0.00 | 4.00 | - | 3.00 | 1.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGTAT.................................................................................................... | 19 | 6.00 | 0.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGTTT.................................................................................................... | 19 | 6.00 | 0.00 | 2.00 | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGAGT.................................................................................................... | 19 | 5.00 | 0.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGACT.................................................................................................... | 19 | 4.00 | 0.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTAAT.................................................................................................... | 19 | 4.00 | 0.00 | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTAGT.................................................................................................... | 19 | 4.00 | 0.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................ACCGATTACATCCCTCGCTCATCCAAT............ | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GTGCAGTTTTCCCATGCAT.................................................................................................... | 19 | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGTGT.................................................................................................... | 19 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTACTC................................................................................................... | 20 | 2.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGAA..................................................................................................... | 18 | 2.00 | 0.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTAATG................................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGGGT.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTCTT.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................ACAGTGAGATGTGCAGAGA.............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGCAC.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTCATCC.................................................................................................. | 21 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTAGTG................................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGTTG.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTAATT................................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTTATA................................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGCAA.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGTCT.................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................CGAAATGTTCTAACTTTTTAAC.......................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................GTGCAGTTTTCCCATGTAG.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTAATC................................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTTGT.................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATCAAT.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................ATCCAATGATTATACCTC. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................GTGCAGTTTTCCCATGGAT.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTCTTC................................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGTTATGACATGCACTACA................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTAA..................................................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATAACT.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGAAG.................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................ATATTATCTTTCTTTAATG................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTGTT.................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGCAG.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGAAA.................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................TGCAGTTTTCCCATTAAT.................................................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................CTACCACATAGTCTTTCTATCTG......................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................GTGCAGTTTTCCCATGTAA.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTGGTC................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTGTTC................................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATTCAT.................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GTGCAGTTTTCCCATGACA.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| TTTTAACTGCCTCTGGAAGTCTATAAACACATGTATAAACAAAAAAGTGAGATATTTTCTACCACATAGTCTTTCTATCTGGTGTCCAAGCTGTATTCTGTTAATTAGTTTGAAAGACTATACAGTGAGATGTGCAGTTTTCCCATTCCATTTTCGAAATGTTCTAACTTTTTAACCTGGTGATATTATCTTTCTTTCAGTTATGACATGCACCGATTACATCCCTCGCTCATCCAATGATTATACCTCA ..............................................................................................................................((((.(((.(((......))).))))))).....................((((.((.........))..)))).................................................. ..............................................................................................................................127......................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR040029(GSM532914) G026T. (cervix) | SRR040043(GSM532928) G428T. (cervix) | SRR040015(GSM532900) G623T. (cervix) | SRR040011(GSM532896) G529T. (cervix) | GSM532876(GSM532876) G547T. (cervix) | SRR040031(GSM532916) G013T. (cervix) | SRR037937(GSM510475) 293cand2. (cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR040025(GSM532910) G613T. (cervix) | SRR189782 | SRR189785 | SRR040039(GSM532924) G531T. (cervix) | SRR040037(GSM532922) G243T. (cervix) | SRR040016(GSM532901) G645N. (cervix) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040022(GSM532907) G575N. (cervix) | TAX577590(Rovira) total RNA. (breast) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR191445(GSM715555) 110genomic small RNA (size selected RNA from . (breast) | TAX577745(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................TTTAACCTGGTGATATCAG............................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................TTTTAACCTGGTGATATG............................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................AACCTGGTGATATTATTA........................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ...........................................................................................................................AGTGAGATGTGCAGTTTTG............................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |