| (3) B-CELL | (9) BREAST | (5) CELL-LINE | (4) CERVIX | (3) HEART | (3) HELA | (4) LIVER | (2) OTHER | (5) SKIN | (1) TESTES | (2) UTERUS | (1) XRN.ip |
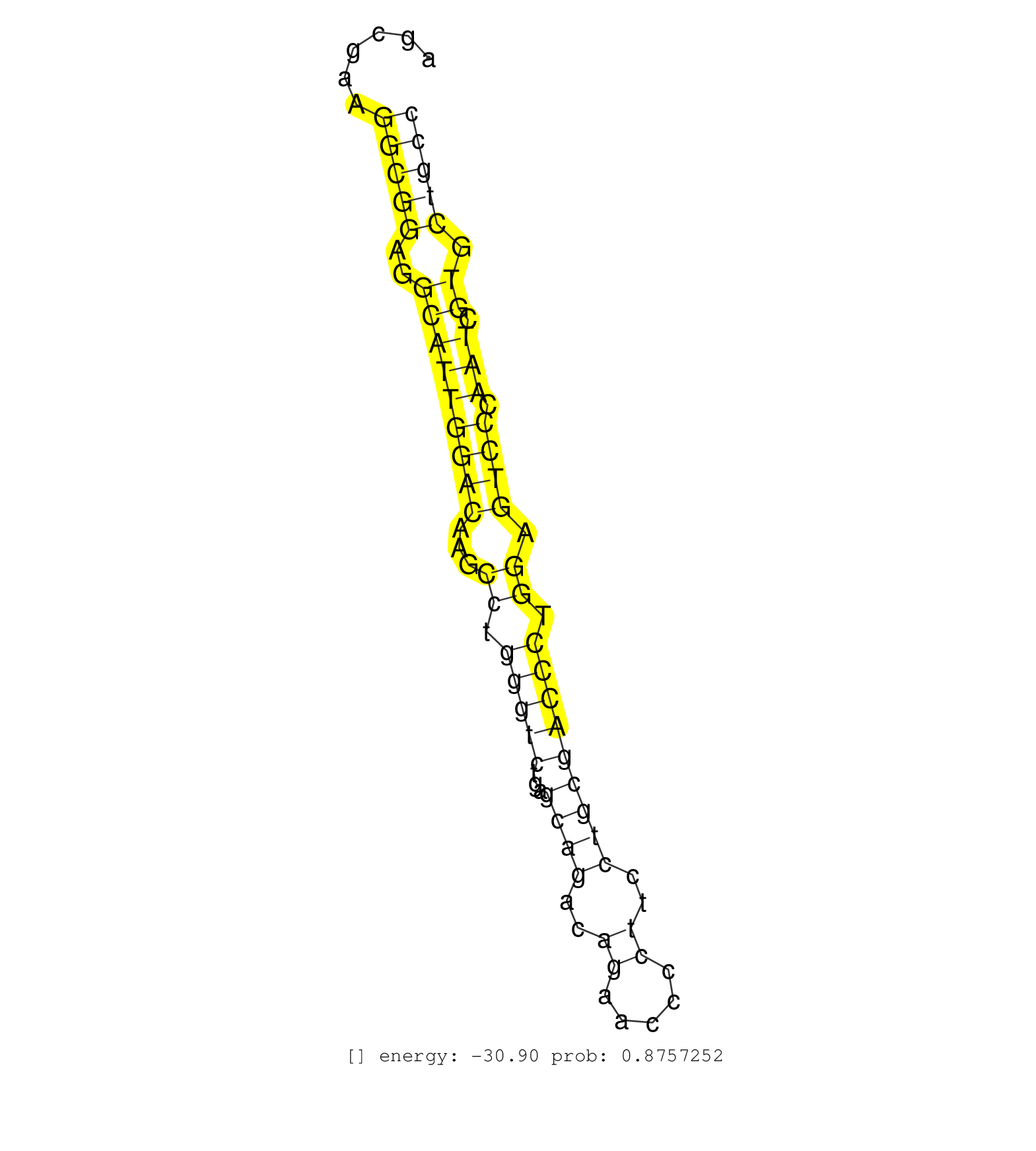
| TATTTCCTGCTTCCTCCCAGCTGACCCTTGAGGGAAAGGGGGTGGGGGCCGCCTAGAGCAGGAGGTATTAGTCACTGCGCCCAGGAGCCAGTTTACACCTAGCGAAGGCGGAGGCATTGGACAAGCCTGGGTCTGAGCAGACAGAACCCCTTCCTGCGACCCTGGAGTCCCAATCGTGCTGCCATTTTCCTTCTGCCCAGGACTCTCCAGTCCTCAGTCACCTTGGACAAAGAAGTGTGGATCCTCAGAT ..........................................................................................................(((((..(((((((((...((.(((((...((((..((.....))..))))))))).)).)))).))).)).)))))................................................................... ....................................................................................................101........................................................................................192........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR343334 | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR189785 | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR038859(GSM458542) MM386. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR343335 | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040010(GSM532895) G529N. (cervix) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | DRR001484(DRX001038) "Hela long nuclear cell fraction, control". (hela) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR191400(GSM715510) 37genomic small RNA (size selected RNA from t. (breast) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | SRR040007(GSM532892) G601T. (cervix) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM532890(GSM532890) G576T. (cervix) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR390723(GSM850202) total small RNA. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359190(GSM359190) HepG2_3pM_4. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR040027(GSM532912) G220T. (cervix) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................AATCGTGCTGCCATTTG.............................................................. | 17 | 1 | 7.00 | 2.00 | 4.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGCGGAGGCATTGGACAAGC............................................................................................................................ | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................AGGCATTGGACAAGCCTGGGTCTGAGCAGACAG.......................................................................................................... | 33 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................AGGCATTGGACAAGCCTGGGTCTGAGCAGAC............................................................................................................ | 31 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AATCGTGCTGCCATTT............................................................... | 16 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGCGGAGGCATTGGACAA.............................................................................................................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CTTGGACAAAGAAGTGTGGA......... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAAGGCGGAGGCATTGGACAA.............................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CGAAGGCGGAGGCATTGGAC................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................GGGTCTGAGCAGACAGAA........................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................CTGAGCAGACAGAACCC..................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CAATCGTGCTGCCATTTG.............................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GCGACCCTGGAGTCCCAATCGTG........................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................AGCGAAGGCGGAGGCATTGGACAAG............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................GGACAAGCCTGGGTCTGAGCAG.............................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATTGGACAAGCCTGGGTC..................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CGAAGGCGGAGGCATTGGACAA.............................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AATCGTGCTGCCATT................................................................ | 15 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CCTTGGACAAAGAAGTGTGGATCCTC.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATTGGACAAGCCTGGGTCT.................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CCTGCGACCCTGGAGTCC................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................ATCGTGCTGCCATTTGA............................................................. | 17 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................CAATCGTGCTGCCATTT............................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGCAGACAGAACCCCTTCCT............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................GCGGAGGCATTGGACAAG............................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................CAAAGAAGTGTGGATCCTCAGA. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................AGGGAAAGGGGGTGGGGG.......................................................................................................................................................................................................... | 18 | 3 | 1.00 | 1.00 | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ...................................................CCTAGAGCAGGAGGTATTAGTCA................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CGAAGGCGGAGGCATTGGACAAG............................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAAGGCGGAGGCATTGGACA............................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGACAAAGAAGTGTGGGGGC...... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................AGGCGGAGGCATTGGACAAGCCTTG........................................................................................................................ | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................ACACCTAGCGAAGGCGGAGGCATTGGACAA.............................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CACCTTGGACAAAGAAGTGTGG.......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................ACCCTGGAGTCCCAATCGTGC....................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TTACACCTAGCGAAGGCGGAG......................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................AGGCATTGGACAAGCCTGG........................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGTCACTGCGCCCAGATG................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ..............................................................................................ACACCTAGCGAAGGCGGAG......................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CGCCCAGGAGCCAGTTTACACCT...................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATTGGACAAGCCTGGGTCTGAGCAGACAG.......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTAGCGAAGGCGGAGGCATTGGAC................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CCAATCGTGCTGCCATT................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TACACCTAGCGAAGGCGGAGGCAT..................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................AGGCATTGGACAAGCCTG......................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CTGGGTCTGAGCAGACAGA......................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................CCTAGCGAAGGCGGAGGCATTGGACAAGCCTGGGTCTGAGCAGACAGAACC...................................................................................................... | 51 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CCAATCGTGCTGCCATTTG.............................................................. | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................AGGCATTGGACAAGCCTGGGTCTGAGCAGA............................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CCCTTCCTGCGACCCGGGT.................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .......................ACCCTTGAGGGAAAGG................................................................................................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .........CTTCCTCCCAGCTGAC................................................................................................................................................................................................................................. | 16 | 9 | 0.22 | 0.22 | - | - | 0.11 | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TATTTCCTGCTTCCTCCCAGCTGACCCTTGAGGGAAAGGGGGTGGGGGCCGCCTAGAGCAGGAGGTATTAGTCACTGCGCCCAGGAGCCAGTTTACACCTAGCGAAGGCGGAGGCATTGGACAAGCCTGGGTCTGAGCAGACAGAACCCCTTCCTGCGACCCTGGAGTCCCAATCGTGCTGCCATTTTCCTTCTGCCCAGGACTCTCCAGTCCTCAGTCACCTTGGACAAAGAAGTGTGGATCCTCAGAT ..........................................................................................................(((((..(((((((((...((.(((((...((((..((.....))..))))))))).)).)))).))).)).)))))................................................................... ....................................................................................................101........................................................................................192........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR343334 | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR189785 | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR038859(GSM458542) MM386. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR343335 | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040010(GSM532895) G529N. (cervix) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | DRR001484(DRX001038) "Hela long nuclear cell fraction, control". (hela) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR191400(GSM715510) 37genomic small RNA (size selected RNA from t. (breast) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | SRR040007(GSM532892) G601T. (cervix) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM532890(GSM532890) G576T. (cervix) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR390723(GSM850202) total small RNA. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359190(GSM359190) HepG2_3pM_4. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR040027(GSM532912) G220T. (cervix) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................CCCTTCCTGCGACCCAT...................................................................................... | 17 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................CAGTCCTCAGTCACCCGGC........................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................CCAGCTGACCCTTGAGCAG....................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |