| (8) B-CELL | (2) BRAIN | (4) BREAST | (33) CELL-LINE | (1) CERVIX | (3) FIBROBLAST | (3) HEART | (2) HELA | (5) LIVER | (2) OTHER | (1) RRP40.ip | (8) SKIN | (1) TESTES | (2) UTERUS |
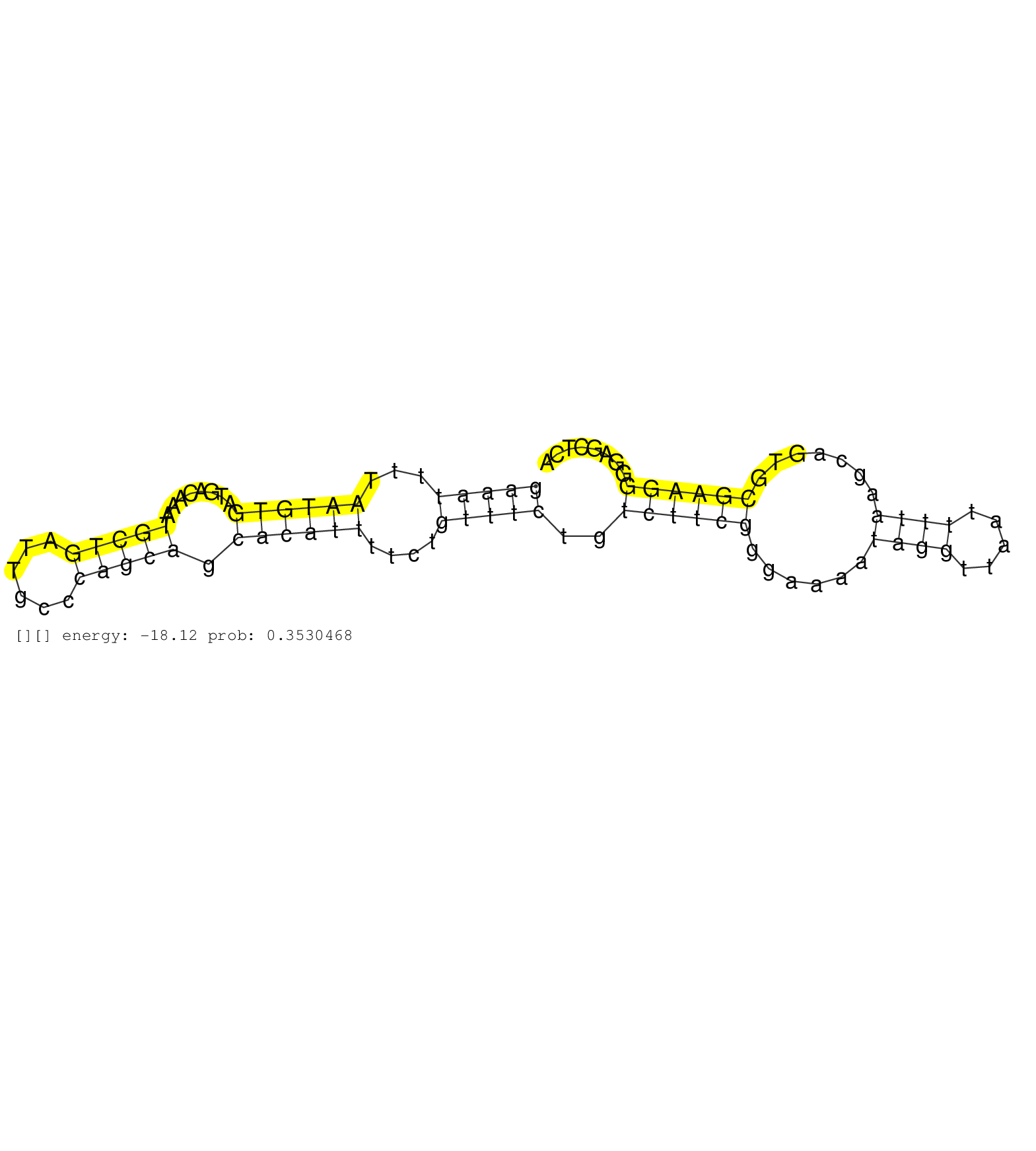
| TTTAAAGTAAATTGATGAACAGCATAACTTAGGGGAATGTGTTTTCTTATGAAATTTTAATGTGATGACAAATGCTGATTGCCCAGCAGCACATTTTCTGTTTCTGTCTTCGGGAAAATAGGTTAATTTTAAGCAGTGCGAAGGGGAGCTCAGGCCAGTGTTTTCACTCCCCGTGTGTCGTTGATGTCTCTCTCCCACAGAACTGAAGCAGATTACCTGGCTCAGTGTCACAGGGCTGCTGATGGTGGTC .............................................................((((.((((...(((((...(((....((((.....((((((...((....)).))))))(((.......))).))))....)))...))).))...)))).))))................................................................................... ..................................................51......................................................................................................................171............................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR029125(GSM416754) U2OS. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR029129(GSM416758) SW480. (cell line) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR189782 | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR029130(GSM416759) DLD2. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191402(GSM715512) 43genomic small RNA (size selected RNA from t. (breast) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | GSM359189(GSM359189) HepG2_2pm_3. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................TAATGTGATGACAAATGCTGATT.......................................................................................................................................................................... | 23 | 1 | 118.00 | 118.00 | - | 27.00 | 16.00 | 12.00 | 10.00 | 3.00 | 7.00 | - | 4.00 | 5.00 | 5.00 | - | 5.00 | - | 4.00 | 2.00 | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - |
| ....................................................................................................................................................................................................ACAGAACTGAAGCAGAATC................................... | 19 | 26.00 | 0.00 | 26.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................TTAATGTGATGACAAATGCTGATT.......................................................................................................................................................................... | 24 | 1 | 26.00 | 26.00 | - | - | 4.00 | - | 2.00 | - | - | - | - | 1.00 | 1.00 | 5.00 | - | - | - | 1.00 | - | - | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................ACAGAACTGAAGCAGAAATC.................................. | 20 | 17.00 | 0.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................CACAGGGCTGCTGATGGTGGT. | 21 | 1 | 6.00 | 6.00 | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TTAATGTGATGACAAATGCTGA............................................................................................................................................................................ | 22 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TAATGTGATGACAAATGCTGATTG......................................................................................................................................................................... | 24 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAACTGAAGCAGATTACCTG............................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................TTAATGTGATGACAAATGCTGATTG......................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TAATGTGATGACAAATGC............................................................................................................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................CAGGGCTGCTGATGGTGGTC | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................AGGGCTGCTGATGGTGGT. | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TCACAGGGCTGCTGATGGTGGT. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TAATGTGATGACAAATGCTGATTGC........................................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AGTGTCACAGGGCTGCTGATGGTGGTCCCCG | 31 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................TTAATGTGATGACAAATGCT.............................................................................................................................................................................. | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAACTGAAGCAGATTACCTGGC............................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TAATGTGATGACAAATGATG............................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................ATTACCTGGCTCAGTGCCAC................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................TTAATGTGATGACAAATG................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................CAGGCCAGTGTTTTCACTCCCCGT............................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................AAGCAGTGCGAAGGGGAGCTCAGGCCAG............................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TCCCCGTGTGTCGTTGATGTCT............................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGAAGCAGATTACCTGGCTCAGTG....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAACTGAAGCAGATTACCTGG.............................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TAATGTGATGACAAATAC............................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................CTCAGTGTCACAGGGCTGCT.......... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................GTCACAGGGCTGCTGATGGTGGT. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GTGCGAAGGGGAGCTCAGGC............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAACTGAAGCAGATTACC................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TTTAAGCAGTGCGAAGGGGAGCTCAGGCCAGCG.......................................................................................... | 33 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................AGTGTCACAGGGCTGCTGATG...... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GTGTCACAGGGCTGCTGATGGT.... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................ACAGGGCTGCTGATGGTGGTTCGG | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................GTCACAGGGCTGCTGATG...... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................CTCTCCCACAGAACTGAAGCAGATTACCTGGC............................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAACTGAAGCAGATTAC.................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TTAATGTGATGACAAAT................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TAATGTGATGACAAATGCTGAT........................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................ACAGGGCTGCTGATGGTTA.. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................ATGTCTCTCTCCCACAGAACTGAAGCAGAC..................................... | 30 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGAACTGAAGCAGATTACCTG............................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TCAGTGTCACAGGGCTGC........... | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AGTGTCACAGGGCTGCTGATGGTGGCC | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................AGGGCTGCTGATGGTGGTCCTC | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ...................................................................................................................................................................................................................................TCACAGGGCTGCTGATGG..... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................ACAGAACTGAAGCAGAAAAA.................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................TAGGGGAATGTGTTTTGTAG......................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................CAGAACTGAAGCAGATT.................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .....................................................................................................................................................................................................................................ACAGGGCTGCTGATGGTG... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTTAAAGTAAATTGATGAACAGCATAACTTAGGGGAATGTGTTTTCTTATGAAATTTTAATGTGATGACAAATGCTGATTGCCCAGCAGCACATTTTCTGTTTCTGTCTTCGGGAAAATAGGTTAATTTTAAGCAGTGCGAAGGGGAGCTCAGGCCAGTGTTTTCACTCCCCGTGTGTCGTTGATGTCTCTCTCCCACAGAACTGAAGCAGATTACCTGGCTCAGTGTCACAGGGCTGCTGATGGTGGTC .............................................................((((.((((...(((((...(((....((((.....((((((...((....)).))))))(((.......))).))))....)))...))).))...)))).))))................................................................................... ..................................................51......................................................................................................................171............................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR029125(GSM416754) U2OS. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR029129(GSM416758) SW480. (cell line) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR189782 | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR029130(GSM416759) DLD2. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191402(GSM715512) 43genomic small RNA (size selected RNA from t. (breast) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | GSM359189(GSM359189) HepG2_2pm_3. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................CTCAGGCCAGTGTTTAGG.................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......GTAAATTGATGAACACTG.................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |