| (1) B-CELL | (2) BRAIN | (8) BREAST | (9) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (7) HEART | (2) HELA | (5) LIVER | (2) OTHER | (11) SKIN | (1) UTERUS |
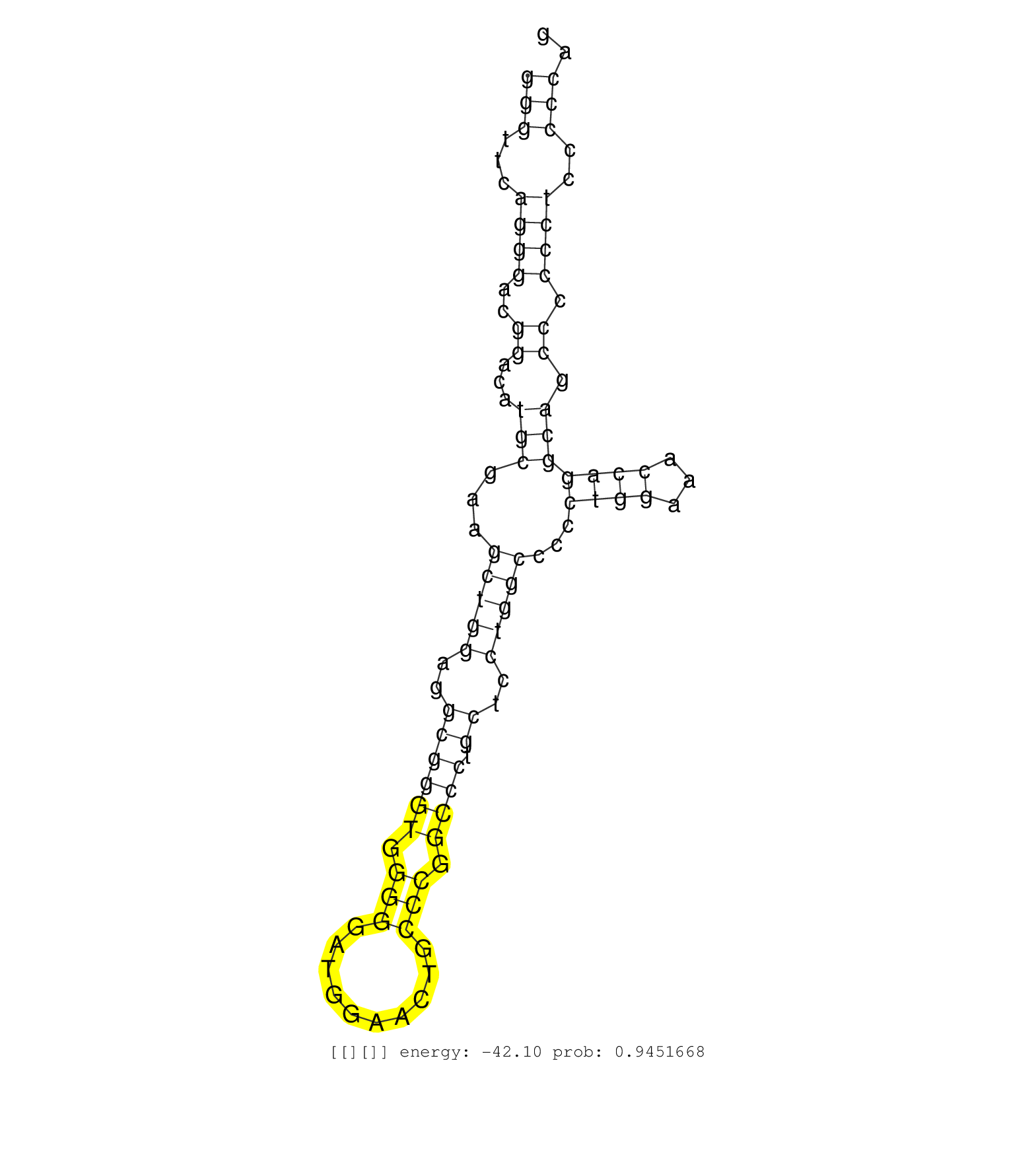
| GCGCCGCCGAGGAGGAGGACGCTATCCACGCCCTCTTCGGCAGCATGCAGGTGAGGGGTGGGGGTTGGGGGATAAACGACCCAGCCGGGACCTGGTGACACAGGGCTGGTCCCTTGCCCTTGTGGATTTCCAGTGTAGTGGGGCAGCAGGCCTTGCACCAAGCACTTTCCTGGAGTTGCTAGGAGAGCTGGGAGGGGAAACCAGGGCCTGCCGCAGGCTGGTGGTGGGGGTGTGAGGTGGTCAGGAAAGACTTCCCTGGGGAGGTGACATTTAAGGTGACGGGACCCAAGCCCTCACCAGGCAGCAGATGCAGGGCTGGTAGGTTGGAGAGATACCTAGTGTAACCAGCAGGTGGCTGGGGTCCGGGGGGTAGGCATGGTGGGAGGTGGCAGGGCTGGGGCGCAAATCAAGAGCATGCTCGCCATCTCTCCCAGGACAGGCTGGCAGAGAGGAAAGCGCTGCTGCTGCAGGCTGTGCAGAGGTG ..............................................................................................................................................................................................................................................................................................................................................(((.......(((((.....))))).......))).......(((.((((((((((.((((....((...........))..)))))))))))))))))................................................... ..............................................................................................................................................................................................................................................................................................................................................335................................................................................................434................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189782 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR040008(GSM532893) G727N. (cervix) | TAX577589(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR029124(GSM416753) HeLa. (hela) | TAX577580(Rovira) total RNA. (breast) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR040007(GSM532892) G601T. (cervix) | TAX577739(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR038860(GSM458543) MM426. (cell line) | SRR189786 | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR191393(GSM715503) 22genomic small RNA (size selected RNA from t. (breast) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | TAX577743(Rovira) total RNA. (breast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCAGT................................................. | 21 | 1 | 17.00 | 1.00 | - | - | 2.00 | 2.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................GGGCTGGGGCGCAAAGCGG.......................................................................... | 19 | 11.00 | 0.00 | - | 5.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................CATGCTCGCCATCTCTCCCAGT................................................. | 22 | 1 | 5.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................CATGCTCGCCATCTCTCCCATT................................................. | 22 | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................TGCTCGCCATCTCTCCCAGT................................................. | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCAGTA................................................ | 22 | 1 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TAGGAGAGCTGGGAGTACA.............................................................................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................................GGTGGTCAGGAAAGAGGGC...................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................................................................................................................................................................TGGTGGGAGGTGGCAGGGCTGGGGCCT................................................................................. | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................GGTGGTGGGGGTGTGGGGT...................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCCGT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................CCCTCTTCGGCAGCACCA.................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .....GCCGAGGAGGAGGACGCTATCCA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCAGTTT............................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................AGGGCTGGTAGGTTGTC............................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCAGTAA............................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................AGCTGGGAGGGGAAATTTT........................................................................................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................................................................................................AGGGCTGGTAGGTTGCCGA.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCTAT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................................................................................................................................................GTGGCTGGGGTCCGGGGGGAAGA.............................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................CATGCTCGCCATCTCTCCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GCCGCCGAGGAGGAGGG................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................AGGCTGGTGGTGGGGGAATG.......................................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ........................................................................................................................................................................................................................................................................................................................................................................................TGGTGGGAGGTGGCAGGGCTGGGGCGCAA............................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................GCATGCTCGCCATCTCTCCCAGA................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................CCTTGTGGATTTCCATTAG............................................................................................................................................................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................GTGGGGGTGTGAGGTGGTCAGGACGCC.......................................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................GGGGGTGTGAGGTGGTTG................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCAG.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................GGAGGTGGCAGGGCTGGGGCAACT............................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCAGA................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................AGGACAGGCTGGCAGGGT.................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................TGGTGGTGGGGGTGTGGGGT...................................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................ATGCTCGCCATCTCTCCCAGC................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................AGAGAGGAAAGCGCTGCTG.................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................GGACAGGCTGGCAGAGA.................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| GCGCCGCCGAGGAGGAGGACGCTATCCACGCCCTCTTCGGCAGCATGCAGGTGAGGGGTGGGGGTTGGGGGATAAACGACCCAGCCGGGACCTGGTGACACAGGGCTGGTCCCTTGCCCTTGTGGATTTCCAGTGTAGTGGGGCAGCAGGCCTTGCACCAAGCACTTTCCTGGAGTTGCTAGGAGAGCTGGGAGGGGAAACCAGGGCCTGCCGCAGGCTGGTGGTGGGGGTGTGAGGTGGTCAGGAAAGACTTCCCTGGGGAGGTGACATTTAAGGTGACGGGACCCAAGCCCTCACCAGGCAGCAGATGCAGGGCTGGTAGGTTGGAGAGATACCTAGTGTAACCAGCAGGTGGCTGGGGTCCGGGGGGTAGGCATGGTGGGAGGTGGCAGGGCTGGGGCGCAAATCAAGAGCATGCTCGCCATCTCTCCCAGGACAGGCTGGCAGAGAGGAAAGCGCTGCTGCTGCAGGCTGTGCAGAGGTG ..............................................................................................................................................................................................................................................................................................................................................(((.......(((((.....))))).......))).......(((.((((((((((.((((....((...........))..)))))))))))))))))................................................... ..............................................................................................................................................................................................................................................................................................................................................335................................................................................................434................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189782 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR040008(GSM532893) G727N. (cervix) | TAX577589(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR029124(GSM416753) HeLa. (hela) | TAX577580(Rovira) total RNA. (breast) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR040007(GSM532892) G601T. (cervix) | TAX577739(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR038860(GSM458543) MM426. (cell line) | SRR189786 | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR191393(GSM715503) 22genomic small RNA (size selected RNA from t. (breast) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | TAX577743(Rovira) total RNA. (breast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................AGGACAGGCTGGCAGAGTATC............................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................TGCTCGCCATCTCTCCCAGAG................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................TTGCACCAAGCACTTAG........................................................................................................................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................GGGCCTGCCGCAGGCTGGTGGTGGGGGTGTCGG........................................................................................................................................................................................................................................................ | 33 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................CCATCTCTCCCAGGACTCGG........................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......CCGAGGAGGAGGACGGCG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................GCTCGCCATCTCTCCTGG.................................................. | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....CGCCGAGGAGGAGGACG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................CACCACCAGCCTGCGG.................................................................................................................................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...CCTCCTCCTCGGCGG.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |