| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (4) AGO2.ip | (11) B-CELL | (11) BRAIN | (14) BREAST | (39) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (4) HEART | (2) HELA | (4) LIVER | (4) OTHER | (31) SKIN | (1) UTERUS | (1) XRN.ip |
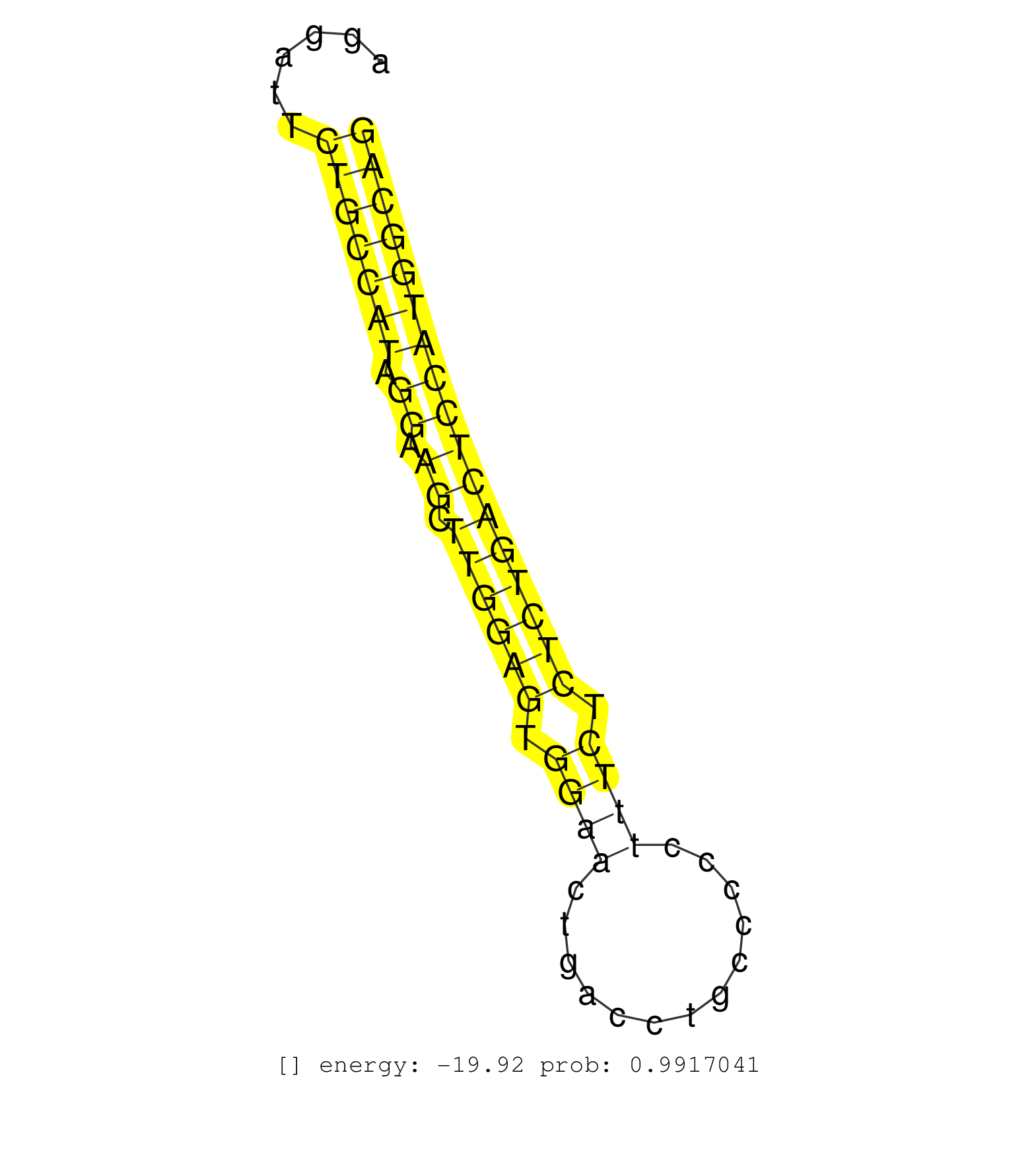
| AAGGCAGCATAGCACTGAGTTCTAGATAACTGAGGGGATTTTGCACTTGGATTTGCTGTGCTAAGGTTTGGGTGGGGCTATTACATCTTGCCGGGCTGGGCAAGAGTGAACCCTAGGGGTCAACATCAGTAGCCAGGATTCTGCCATAGGAAGCTTGGAGTGGAACTGACCTGCCCCCTTTCTCTCTGACTCCATGGCAGCTGGGTGAGCTATGAACAGGCCAACTGCAAGGGCGAGCAGTTTGTGTTTG ............................................................................................................................................(((((((.((.((.((((((.((((.............)))).))))))))))))))))).................................................. ......................................................................................................................................135..............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038863(GSM458546) MM603. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR390723(GSM850202) total small RNA. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037943(GSM510481) 293DcrTN. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040013(GSM532898) G648T. (cervix) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR191604(GSM715714) 74genomic small RNA (size selected RNA from t. (breast) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191566(GSM715676) 94genomic small RNA (size selected RNA from t. (breast) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR191454(GSM715564) 180genomic small RNA (size selected RNA from . (breast) | SRR191630(GSM715740) 70genomic small RNA (size selected RNA from t. (breast) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR015358(GSM380323) Na╠°ve B Cell (Naive39). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR038860(GSM458543) MM426. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR038857(GSM458540) D20. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR040024(GSM532909) G613N. (cervix) | SRR038862(GSM458545) MM472. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191618(GSM715728) 148genomic small RNA (size selected RNA from . (breast) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATT................................................. | 21 | 1 | 62.00 | 9.00 | - | 5.00 | 5.00 | 5.00 | 3.00 | - | 1.00 | - | 1.00 | 3.00 | - | - | 4.00 | 3.00 | 2.00 | 3.00 | 3.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | 2.00 | 1.00 | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGT................................................. | 21 | 1 | 53.00 | 10.00 | 16.00 | - | 2.00 | - | 1.00 | 3.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGA................................................. | 21 | 1 | 36.00 | 10.00 | 13.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | 2.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAT.................................................. | 20 | 1 | 26.00 | 9.00 | 1.00 | 10.00 | 1.00 | 4.00 | 1.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAA.................................................. | 20 | 1 | 18.00 | 9.00 | 6.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAAA................................................. | 21 | 1 | 13.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAAT................................................. | 21 | 1 | 10.00 | 9.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TCTGCCATAGGAAGCTTGGAGTGG....................................................................................... | 24 | 2 | 10.00 | 10.00 | - | - | - | - | 0.50 | 1.50 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAG.................................................. | 20 | 1 | 10.00 | 10.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCA................................................... | 19 | 1 | 9.00 | 9.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATTT................................................ | 22 | 1 | 7.00 | 9.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAATT................................................ | 22 | 1 | 6.00 | 9.00 | 1.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGTATC.............................................. | 24 | 1 | 6.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGTT................................................ | 22 | 1 | 5.00 | 10.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGC................................................. | 21 | 1 | 5.00 | 5.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTCTCTCTGACTCCATGGCAGA................................................. | 22 | 1 | 4.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAAAA................................................ | 22 | 1 | 4.00 | 9.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGTA................................................ | 22 | 1 | 4.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATA................................................. | 21 | 1 | 3.00 | 9.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CTTGGAGTGGAACTGACCTGA............................................................................ | 21 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TTCTCTCTGACTCCATGGCA................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATTA................................................ | 22 | 1 | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGCAA............................................... | 23 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATAA................................................ | 22 | 1 | 2.00 | 9.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAATAA............................................... | 23 | 1 | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTCTCTCTGACTCCATGGCAGT................................................. | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTCTCTCTGACTCCATGGCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCTGCCATAGGAAGCTTGGAG.......................................................................................... | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - |
| ...................................................................................................................................................................................TTCTCTCTGACTCCATGGCAAT................................................. | 22 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGAT................................................ | 22 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTCTCTCTGACTCCATGGCATTT................................................ | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TCTGCCATAGGAAGCTTGGAGCGGA...................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGGAA............................................... | 23 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTCTCTCTGACTCCATGGCAT.................................................. | 21 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGGGTGAGCTATGAACAGGCC............................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATTTC............................................... | 23 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCCGA................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAAC................................................. | 21 | 1 | 1.00 | 9.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCGTTT................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCCGC................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATCA................................................ | 22 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTTCTCTCTGACTCCATGGCATT................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................CCTTTCTCTCTGACTCCATGGCAGA................................................. | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAAG................................................. | 21 | 1 | 1.00 | 9.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATAT................................................ | 22 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGG................................................. | 21 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TCTGCCATAGGAAGCTTGGAGTGA....................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................CCCCCTTTCTCTCTGACTCCATGGA.................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCATC................................................. | 21 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAATA................................................ | 22 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCCGTA................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCACT................................................. | 21 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGGCAGCA................................................ | 22 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTCTGACTCCATGGCAGATT............................................... | 21 | 3 | 0.67 | 0.33 | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTCTGACTCCATGGCAGAAA............................................... | 21 | 3 | 0.67 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CCGGGCTGGGCAAGAGTGAACCCTAGGGGTCAAC.............................................................................................................................. | 34 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................AGATAACTGAGGGGATTTTGCACTTGG........................................................................................................................................................................................................ | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ...........................................................................................................................................TCTGCCATAGGAAGCTTGGAGTGGA...................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGCCATAGGAAGCTTGGAGTGG....................................................................................... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TCTGCCATAGGAAGCTTGGA........................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GCTGGGCAAGAGTGAACCCTAGGGGTCA................................................................................................................................ | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ............................................................................................................................................CTGCCATAGGAAGCTTGGAGTGG....................................................................................... | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTCTCTGACTCCATGG..................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTCTGACTCCATGGCAGAATT.............................................. | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ......................................................................................................................................................................................TCTCTGACTCCATGGCAG.................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTCTGACTCCATGGCAGAACT.............................................. | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TGACCTGCCCCCTTTCT................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| ........................................................................................................................................................GCTTGGAGTGGAACTCTG................................................................................ | 18 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| ........................................................................................................................................................GCTTGGAGTGGAACT................................................................................... | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................GGTGAGCTATGAACAG............................... | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GTTTGGGTGGGGCTA.......................................................................................................................................................................... | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGGCAGCATAGCACTGAGTTCTAGATAACTGAGGGGATTTTGCACTTGGATTTGCTGTGCTAAGGTTTGGGTGGGGCTATTACATCTTGCCGGGCTGGGCAAGAGTGAACCCTAGGGGTCAACATCAGTAGCCAGGATTCTGCCATAGGAAGCTTGGAGTGGAACTGACCTGCCCCCTTTCTCTCTGACTCCATGGCAGCTGGGTGAGCTATGAACAGGCCAACTGCAAGGGCGAGCAGTTTGTGTTTG ............................................................................................................................................(((((((.((.((.((((((.((((.............)))).))))))))))))))))).................................................. ......................................................................................................................................135..............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038863(GSM458546) MM603. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR390723(GSM850202) total small RNA. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037943(GSM510481) 293DcrTN. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040013(GSM532898) G648T. (cervix) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR191604(GSM715714) 74genomic small RNA (size selected RNA from t. (breast) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191566(GSM715676) 94genomic small RNA (size selected RNA from t. (breast) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR191454(GSM715564) 180genomic small RNA (size selected RNA from . (breast) | SRR191630(GSM715740) 70genomic small RNA (size selected RNA from t. (breast) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR015358(GSM380323) Na╠°ve B Cell (Naive39). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR038860(GSM458543) MM426. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR038857(GSM458540) D20. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR040024(GSM532909) G613N. (cervix) | SRR038862(GSM458545) MM472. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191618(GSM715728) 148genomic small RNA (size selected RNA from . (breast) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................ACCCAAACCTTAGCACAGC................................................................................................................................................................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |