| (1) AGO1.ip | (3) B-CELL | (11) BRAIN | (6) CELL-LINE | (1) HEART | (2) OTHER | (1) SKIN | (1) UTERUS |
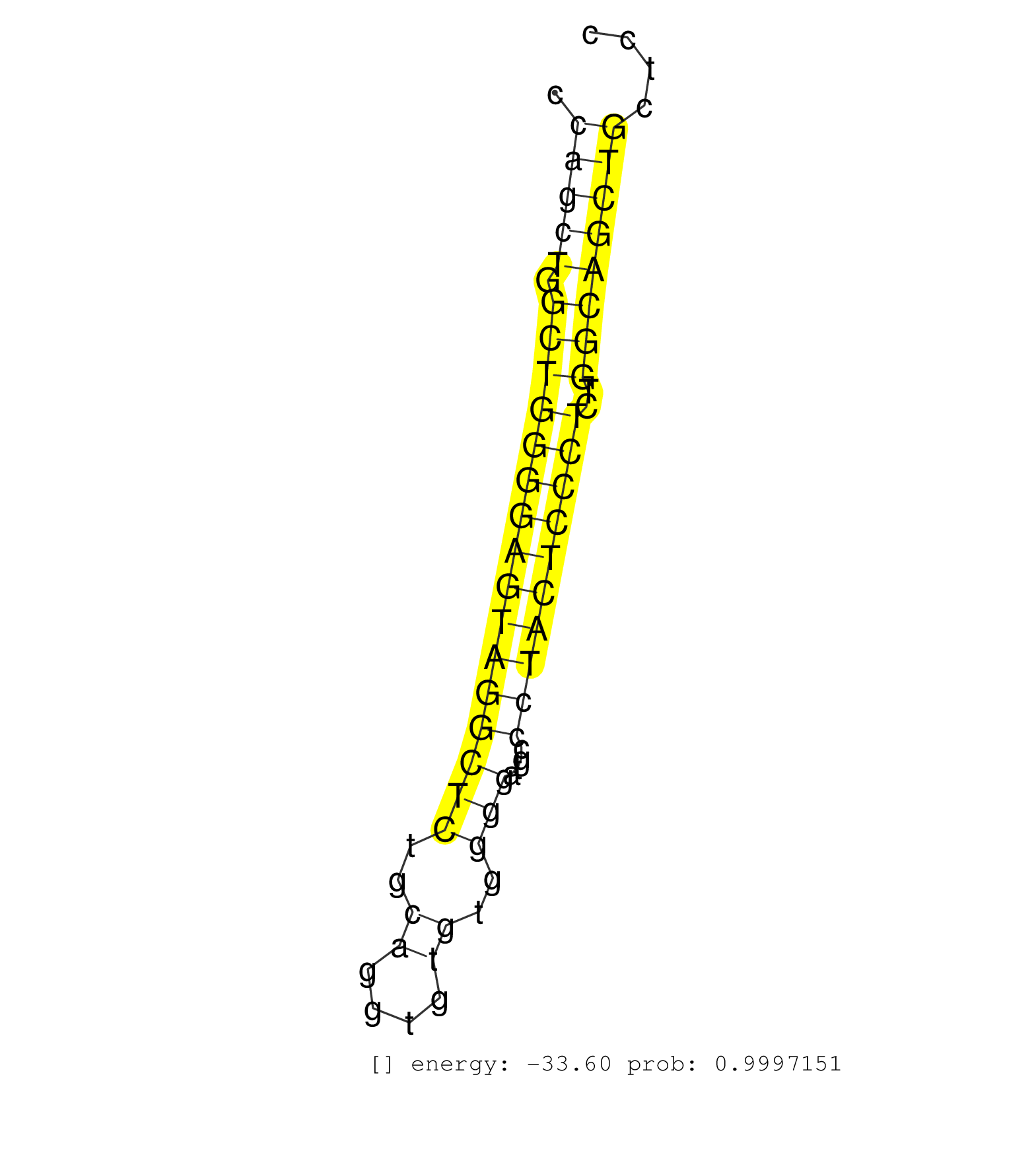
| CTGCACCTGGGCAGACCCTGAACCTACCTGTCATGGGCGTTGTTGTCCAGGTGAGAGCCAGCTGGCTGGGGAGTAGGCTCTGCAGGTGTGTGGGGATGCCCTACTCCCTCTGGCAGCTGCTCCCTCCTTCTCCCAGGTGCGCATCCCATCCAGGGTGGACAAGTCCGAGTCCAGTCCTCCGAAGCA ..........................................................(((((.((((((((((((((((..((....))..)))....))))))))))..))))))))................................................................... .........................................................58...............................................................123............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................TGGCTGGGGAGTAGGCTC.......................................................................................................... | 18 | 1 | 10.00 | 10.00 | - | - | 4.00 | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................TGGCTGGGGAGTAGGCTCTGC....................................................................................................... | 21 | 1 | 10.00 | 10.00 | 4.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGCTGGGGAGTAGGCTCT......................................................................................................... | 19 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGCTGGGGAGTAGGCTCTGCA...................................................................................................... | 22 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................AGCCAGCTGGCTGGGGAGTAG.............................................................................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGCTGGGGAGTAGGCTC.......................................................................................................... | 17 | 2 | 2.50 | 2.50 | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ..............................................................TGGCTGGGGAGTAGGCTCTGCAG..................................................................................................... | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGCTGGGGAGTAGGCT........................................................................................................... | 17 | 3 | 1.67 | 1.67 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | 0.33 | - | 0.33 | 0.33 |
| .....................................................................................................TACTCCCTCTGGCAGCTG................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TACTCCCTCTGGCAGCTGCTTA............................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................GGCTGGGGAGTAGGCAA.......................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................TACTCCCTCTGGCAGCTGT.................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TACTCCCTCTGGCAGCTGTTTT............................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TACTCCCTCTGGCAGCTGCTCC............................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ACTCCCTCTGGCAGCTGTAA................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .............................................................CTGGCTGGGGAGTAGGCT........................................................................................................... | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGCTGGGGAGTAGGCTCTGCAGG.................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................GCTGGGGAGTAGGCTCTGC....................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GCTCTGCAGGTGTGTGGGGATG........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AGGTGCGCATCCCATACCT................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ....................................................GAGAGCCAGCTGGCTGGGGAGTAGGC............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................GGCTGGGGAGTAGGCTCT......................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| CTGCACCTGGGCAGACCCTGAACCTACCTGTCATGGGCGTTGTTGTCCAGGTGAGAGCCAGCTGGCTGGGGAGTAGGCTCTGCAGGTGTGTGGGGATGCCCTACTCCCTCTGGCAGCTGCTCCCTCCTTCTCCCAGGTGCGCATCCCATCCAGGGTGGACAAGTCCGAGTCCAGTCCTCCGAAGCA ..........................................................(((((.((((((((((((((((..((....))..)))....))))))))))..))))))))................................................................... .........................................................58...............................................................123............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) |
|---|