| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (2) AGO2.ip | (2) B-CELL | (2) BRAIN | (4) BREAST | (23) CELL-LINE | (1) CERVIX | (1) HEART | (2) HELA | (3) LIVER | (2) OTHER | (1) RRP40.ip | (5) SKIN | (1) XRN.ip |
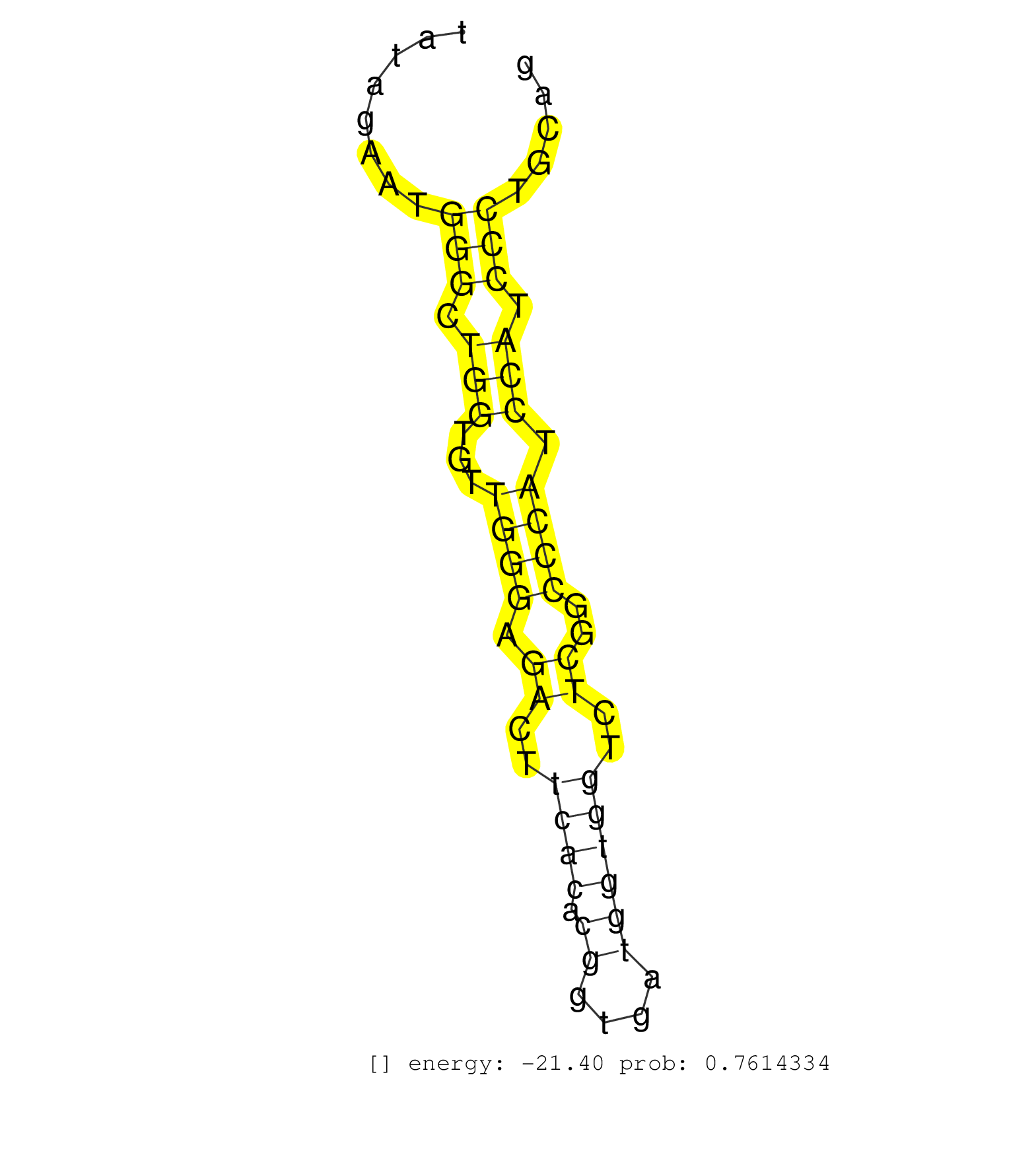
| AGCAGTGCTGGGAAATGTGTACAGATGAGGAAACTGAGGCACCGAGAGGGCAGTGGTTCAGAGTCCATGGCCCCTGACTGCTCCCCAGCCCGCCTTTCCAGGGGCCTGGCCTCACTGCGGCAGCGTCCCCGGCTATAGAATGGGCTGGTGTTGGGAGACTTCACACGGTGATGGTGGTCTCGGCCCATCCATCCCTGCAGCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGC .............................................................................................................................................(((.(((...((((.((..((((.((....))))))..))..)))).))).)))....................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR038858(GSM458541) MEL202. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191559(GSM715669) 65genomic small RNA (size selected RNA from t. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGACT.......................................................................................... | 22 | 1 | 13.00 | 13.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................TCTCGGCCCATCCATCCCTGC.................................................... | 21 | 1 | 8.00 | 8.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGACTT......................................................................................... | 23 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGACTTC........................................................................................ | 24 | 1 | 6.00 | 6.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGAC........................................................................................... | 21 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGACTTTAA...................................................................................... | 26 | 1 | 2.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GGTGTTGGGAGACTTCACACGG.................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CCATCCATCCCTGCAATAC............................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGACTAA........................................................................................ | 24 | 1 | 2.00 | 13.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CAAGACGTGCTCCCAGGACGAGTTT..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CCCGGCTATAGAATGGGC......................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GAATGGGCTGGTGTTGGGAGACTTC........................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGAGTCCATGGCCCCTGACT........................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TCTCGGCCCATCCATCCCTGCAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................GTCCCCGGCTATAGAATGGG.......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GAGGAAACTGAGGCACCGAGAGGGC....................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GGAAATGTGTACAGATGAGGAAACTGAG.................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCGGCTATAGAATGGG.......................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ATGGGCTGGTGTTGGGAGACTTC........................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CTGACTGCTCCCCAGCCAA.............................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................GTTTCGCTGCCACGATGGGAAGT.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CCATCCATCCCTGCAATAT............................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGAGTCCATGGCCCCTGACA........................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................AAGACGTGCTCCCAGGACGAA........................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................ATGAGGAAACTGAGGCACCGAGAGGTT....................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....TGCTGGGAAATGTGTACAGATGAGGAAA......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGCCCCCAAGACGTGCTCCCAG.............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TATAGAATGGGCTGGTGTTG................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................................GTTTCGCTGCCACGATGGGAAG... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGAGGAAACTGAGGCACCGAGAGGGCC...................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................CGAGAGGGCAGTGGTTCAGAG........................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGACA.......................................................................................... | 22 | 1 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CCCCAAGACGTGCTCCCAGGACGAGTTT..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAG............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................GTGTACAGATGAGGAAACTGAGGC.................................................................................................................................................................................................................. | 24 | 3 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AATGGGCTGGTGTTGGGAGACTTA........................................................................................ | 24 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GAGGAAACTGAGGCACCGAGAG.......................................................................................................................................................................................................... | 22 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CTGACTGCTCCCCAGC................................................................................................................................................................. | 16 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................CCTGACTGCTCCCCAGC................................................................................................................................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GAGGAAACTGAGGCACCGAGA........................................................................................................................................................................................................... | 21 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| AGCAGTGCTGGGAAATGTGTACAGATGAGGAAACTGAGGCACCGAGAGGGCAGTGGTTCAGAGTCCATGGCCCCTGACTGCTCCCCAGCCCGCCTTTCCAGGGGCCTGGCCTCACTGCGGCAGCGTCCCCGGCTATAGAATGGGCTGGTGTTGGGAGACTTCACACGGTGATGGTGGTCTCGGCCCATCCATCCCTGCAGCCCCCAAGACGTGCTCCCAGGACGAGTTTCGCTGCCACGATGGGAAGTGC .............................................................................................................................................(((.(((...((((.((..((((.((....))))))..))..)))).))).)))....................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR038858(GSM458541) MEL202. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM359202(GSM359202) hepg2_untreated_a. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191559(GSM715669) 65genomic small RNA (size selected RNA from t. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................GTTCAGAGTCCATGGTCTG................................................................................................................................................................................ | 19 | 4.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................CCCCAAGACGTGCTCCCACAC............................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................GTTCAGAGTCCATGGCTCTG............................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................CAAGACGTGCTCCCATA............................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .................TGTACAGATGAGGAAACTGAGGCAG................................................................................................................................................................................................................ | 25 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................CCAAGACGTGCTCCCAAA............................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .................................................................................TCCCCAGCCCGCCTTT......................................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................GATGGTGGTCTCGG................................................................... | 14 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |