| (1) AGO1.ip | (4) AGO2.ip | (2) AGO3.ip | (6) B-CELL | (2) BRAIN | (6) BREAST | (22) CELL-LINE | (1) FIBROBLAST | (2) HEART | (3) HELA | (1) KIDNEY | (4) LIVER | (2) OTHER | (17) SKIN | (1) TESTES | (1) XRN.ip |
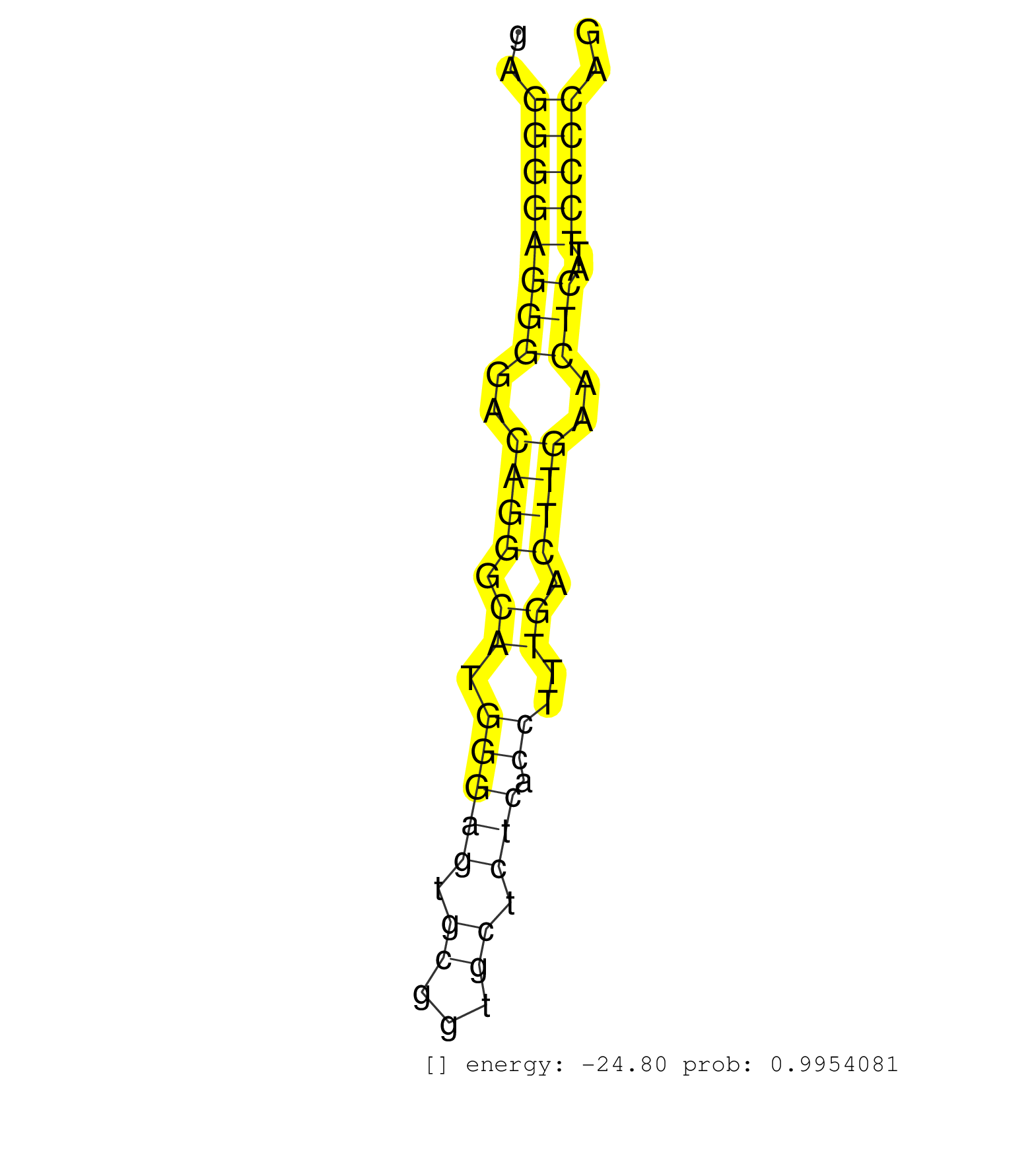
| GCCCAGCACTCTCAGTGGGGAGCCAGGTGGGAGAACAGGCTCGGAAGGGGACCTAGGCTTATGCAGCGAGCCGGGCAAAGCTGGAACTGGAGCCCAGGCCCCTGGATGCCCCCTGGCTTGTGGAGTTCTGGGATACTGAGGGGAGGGGACAGGGCATGGGAGTGCGGTGCTCTCACCTTTGACTTGAACTCATTCCCCAGGGGACAGGGGAGGCCTCCTCAGGATCCACAGATGCCCAGTCTCCCAAGAG ...........................................................................................................................................((((((((..((((.((.(((((.((...)).))).))..)).))))..)))..))))).................................................... .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189783 | SRR189782 | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189784 | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | TAX577579(Rovira) total RNA. (breast) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | TAX577588(Rovira) total RNA. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | TAX577741(Rovira) total RNA. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR189787 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAGT................................................. | 24 | 1 | 42.00 | 14.00 | 4.00 | 1.00 | 2.00 | 4.00 | 1.00 | 2.00 | 2.00 | 3.00 | - | 3.00 | 2.00 | - | 1.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAGT................................................. | 23 | 1 | 16.00 | 10.00 | - | 1.00 | 4.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAG.................................................. | 23 | 1 | 14.00 | 14.00 | - | 2.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAG.................................................. | 22 | 1 | 10.00 | 10.00 | 7.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGGGACAGGGGAGGGCTA................................. | 20 | 8.00 | 0.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAGA................................................. | 23 | 1 | 8.00 | 10.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAGAA................................................ | 24 | 1 | 5.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAGA................................................. | 24 | 1 | 5.00 | 14.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGGGACAGGGGAGGGC................................... | 18 | 4.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................GAGAACAGGCTCGGAAG........................................................................................................................................................................................................... | 17 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................GCTCGGAAGGGGACCTAGGCTTATGCA......................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAGTA................................................ | 24 | 1 | 2.00 | 10.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAT.................................................. | 23 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAGG................................................. | 23 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TCACCTTTGACTTGATCC............................................................ | 18 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAA.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAAG................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGGGACAGGGGAGGCCTA................................. | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................TGCGGTGCTCTCACCGCCG..................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAGAGT............................................... | 25 | 1 | 1.00 | 10.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCC.................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GCTTGTGGAGTTCTGCGG..................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................GTGCGGTGCTCTCACATTA...................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAGAAC............................................... | 26 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAAT................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GAGGGGAGGGGACAGGGCAGGGG.......................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCC.................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAGTAG............................................... | 25 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGGGAGGGGACAGGGCATGGG.......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GGGACAGGGCATGGGAGGCAG.................................................................................... | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCAT.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACTTGAACTCATTCCCCAGAGAA.............................................. | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGACTTGAACTCATTCCCCATT................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAA.................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAGC................................................. | 24 | 1 | 1.00 | 14.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCCAGTA................................................ | 25 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTTGACTTGAACTCATTCCCTAA.................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................CAGGGGACAGGGGAGGC.................................... | 17 | 6 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.17 |
| GCCCAGCACTCTCAGTGGGGAGCCAGGTGGGAGAACAGGCTCGGAAGGGGACCTAGGCTTATGCAGCGAGCCGGGCAAAGCTGGAACTGGAGCCCAGGCCCCTGGATGCCCCCTGGCTTGTGGAGTTCTGGGATACTGAGGGGAGGGGACAGGGCATGGGAGTGCGGTGCTCTCACCTTTGACTTGAACTCATTCCCCAGGGGACAGGGGAGGCCTCCTCAGGATCCACAGATGCCCAGTCTCCCAAGAG ...........................................................................................................................................((((((((..((((.((.(((((.((...)).))).))..)).))))..)))..))))).................................................... .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189783 | SRR189782 | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189784 | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | TAX577579(Rovira) total RNA. (breast) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | TAX577588(Rovira) total RNA. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | TAX577741(Rovira) total RNA. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR189787 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................GGGAGAACAGGCTCGGATCTT......................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................GAGTTCTGGGATACTGTG.............................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| .........................................................................................................................................................................................GAACTCATTCCCCAGTGAT.............................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |